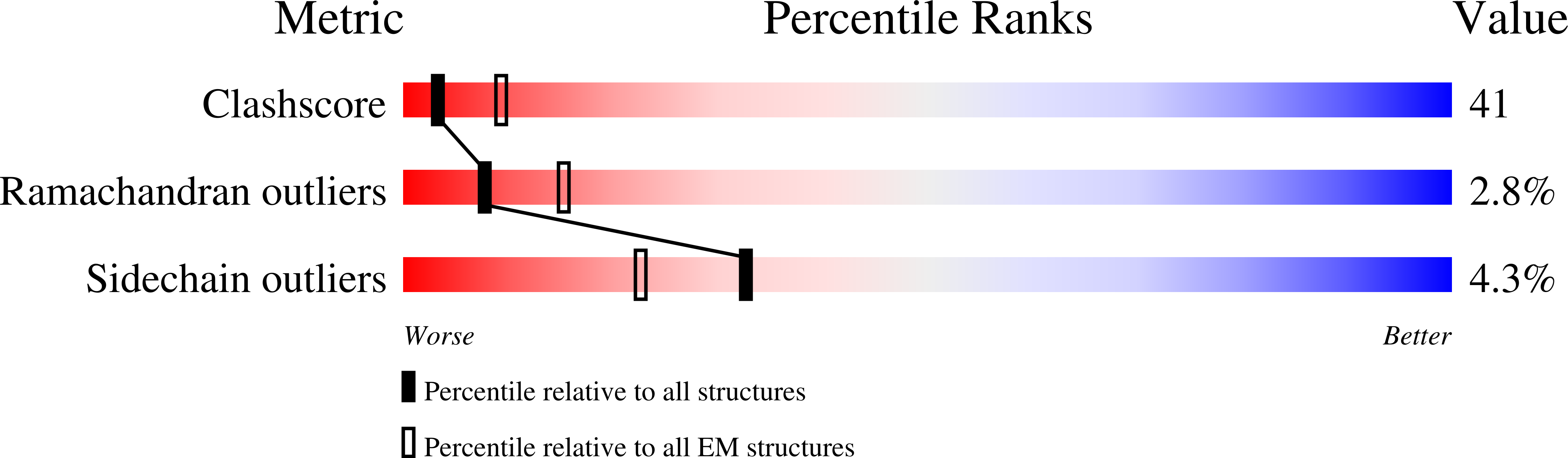Multi-modal adaptor-clathrin contacts drive coated vesicle assembly.
Smith, S.M., Larocque, G., Wood, K.M., Morris, K.L., Roseman, A.M., Sessions, R.B., Royle, S.J., Smith, C.J.(2021) EMBO J 40: e108795-e108795
- PubMed: 34487371
- DOI: https://doi.org/10.15252/embj.2021108795
- Primary Citation of Related Structures:
7OM8 - PubMed Abstract:
Clathrin-coated pits are formed by the recognition of membrane and cargo by the AP2 complex and the subsequent recruitment of clathrin triskelia. A role for AP2 in coated-pit assembly beyond initial clathrin recruitment has not been explored. Clathrin binds the β2 subunit of AP2, and several binding sites have been identified, but our structural knowledge of these interactions is incomplete and their functional importance during endocytosis is unclear. Here, we analysed the cryo-EM structure of clathrin cages assembled in the presence of β2 hinge-appendage (β2HA). We find that the β2-appendage binds in at least two positions in the cage, demonstrating that multi-modal binding is a fundamental property of clathrin-AP2 interactions. In one position, β2-appendage cross-links two adjacent terminal domains from different triskelia. Functional analysis of β2HA-clathrin interactions reveals that endocytosis requires two clathrin interaction sites: a clathrin-box motif on the hinge and the "sandwich site" on the appendage. We propose that β2-appendage binding to more than one triskelion is a key feature of the system and likely explains why assembly is driven by AP2.
Organizational Affiliation:
School of Life Sciences, University of Warwick, Coventry, UK.