Covalent flexible peptide docking in Rosetta.
Tivon, B., Gabizon, R., Somsen, B.A., Cossar, P.J., Ottmann, C., London, N.(2021) Chem Sci 12: 10836-10847
- PubMed: 34476063
- DOI: https://doi.org/10.1039/d1sc02322e
- Primary Citation of Related Structures:
7O07 - PubMed Abstract:
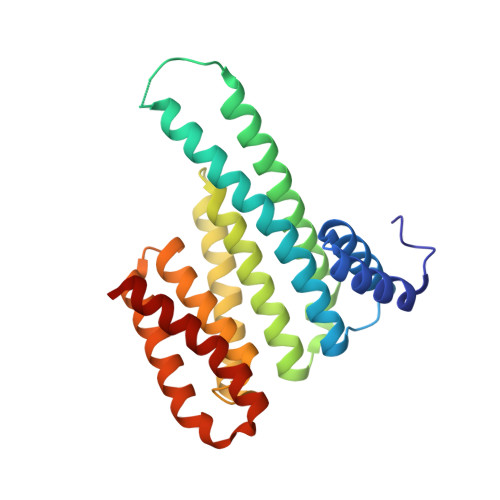
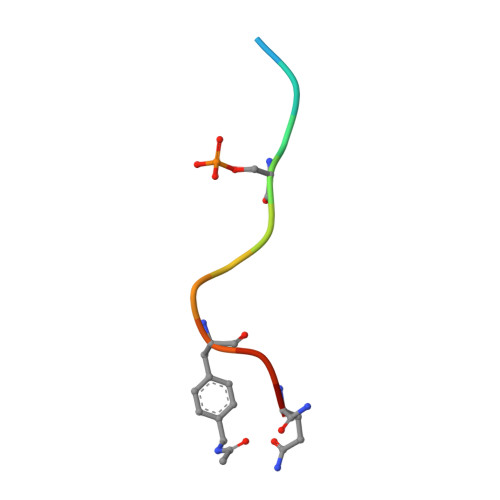
Electrophilic peptides that form an irreversible covalent bond with their target have great potential for binding targets that have been previously considered undruggable. However, the discovery of such peptides remains a challenge. Here, we present Rosetta CovPepDock, a computational pipeline for peptide docking that incorporates covalent binding between the peptide and a receptor cysteine. We applied CovPepDock retrospectively to a dataset of 115 disulfide-bound peptides and a dataset of 54 electrophilic peptides. It produced a top-five scoring, near-native model, in 89% and 100% of the cases when docking from the native conformation, and 20% and 90% when docking from an extended peptide conformation, respectively. In addition, we developed a protocol for designing electrophilic peptide binders based on known non-covalent binders or protein-protein interfaces. We identified 7154 peptide candidates in the PDB for application of this protocol. As a proof-of-concept we validated the protocol on the non-covalent complex of 14-3-3σ and YAP1 phosphopeptide. The protocol identified seven highly potent and selective irreversible peptide binders. The predicted binding mode of one of the peptides was validated using X-ray crystallography. This case-study demonstrates the utility and impact of CovPepDock. It suggests that many new electrophilic peptide binders can be rapidly discovered, with significant potential as therapeutic molecules and chemical probes.
Organizational Affiliation:
Department of Chemical and Structural Biology, The Weizmann Institute of Science Rehovot 7610001 Israel nir.london@weizmann.ac.il.