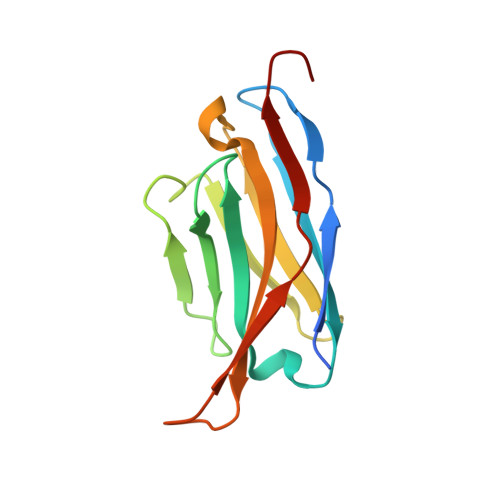
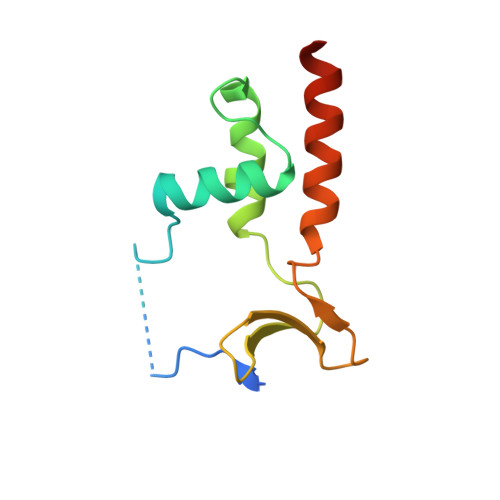
Molecular mechanisms underlying the role of the centriolar CEP164-TTBK2 complex in ciliopathies.
Rosa E Silva, I., Bino, L., Johnson, C.M., Rutherford, T.J., Neuhaus, D., Andreeva, A., Cajanek, L., van Breugel, M.(2022) Structure 30: 114-128.e9
- PubMed: 34499853
- DOI: https://doi.org/10.1016/j.str.2021.08.007
- Primary Citation of Related Structures:
7NWJ, 7O06, 7O0S, 7O3B - PubMed Abstract:
Cilia formation is essential for human life. One of the earliest events in the ciliogenesis program is the recruitment of tau-tubulin kinase 2 (TTBK2) by the centriole distal appendage component CEP164. Due to the lack of high-resolution structural information on this complex, it is unclear how it is affected in human ciliopathies such as nephronophthisis. Furthermore, it is poorly understood if binding to CEP164 influences TTBK2 activities. Here, we present a detailed biochemical, structural, and functional analysis of the CEP164-TTBK2 complex and demonstrate how it is compromised by two ciliopathic mutations in CEP164. Moreover, we also provide insights into how binding to CEP164 is coordinated with TTBK2 activities. Together, our data deepen our understanding of a crucial step in cilia formation and will inform future studies aimed at restoring CEP164 functionality in a debilitating human ciliopathy.
Organizational Affiliation:
Queen Mary University of London, School of Biological and Chemical Sciences, 2 Newark Street, London E1 2AT, UK; Medical Research Council - Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK. Electronic address: ivan.silva@alumni.usp.br.