Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222.
Smialek, J., Nowakowski, M., Bzowska, M., Bochenska, O., Wlizlo, A., Kozik, A., Dubin, G., Mak, P.(2021) Int J Mol Sci 22
- PubMed: 34200765
- DOI: https://doi.org/10.3390/ijms22126256
- Primary Citation of Related Structures:
7NYI - PubMed Abstract:
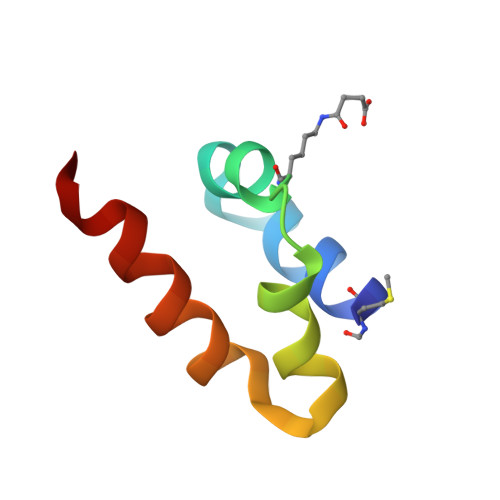
BacSp222 is a multifunctional peptide produced by Staphylococcus pseudintermedius 222. This 50-amino acid long peptide belongs to subclass IId of bacteriocins and forms a four-helix bundle molecule. In addition to bactericidal functions, BacSp222 possesses also features of a virulence factor, manifested in immunomodulatory and cytotoxic activities toward eukaryotic cells. In the present study, we demonstrate that BacSp222 is produced in several post-translationally modified forms, succinylated at the ε-amino group of lysine residues. Such modifications have not been previously described for any bacteriocins. NMR and circular dichroism spectroscopy studies have shown that the modifications do not alter the spatial structure of the peptide. At the same time, succinylation significantly diminishes its bactericidal and cytotoxic potential. We demonstrate that the modification of the bacteriocin is an effect of non-enzymatic reaction with a highly reactive intracellular metabolite, i.e., succinyl-coenzyme A. The production of succinylated forms of the bacteriocin depends on environmental factors and on the access of bacteria to nutrients. Our study indicates that the production of succinylated forms of bacteriocin occurs in response to the changing environment, protects producer cells against the autotoxicity of the excreted peptide, and limits the pathogenicity of the strain.
Organizational Affiliation:
Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7 St., 30-387 Kraków, Poland.