Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
Barlaam, B., De Savi, C., Dishington, A., Drew, L., Ferguson, A.D., Ferguson, D., Gu, C., Hande, S., Hassall, L., Hawkins, J., Hird, A.W., Holmes, J., Lamb, M.L., Lister, A.S., McGuire, T.M., Moore, J.E., O'Connell, N., Patel, A., Pike, K.G., Sarkar, U., Shao, W., Stead, D., Varnes, J.G., Vasbinder, M.M., Wang, L., Wu, L., Xue, L., Yang, B., Yao, T.(2021) J Med Chem 64: 15189-15213
- PubMed: 34647738
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01249
- Primary Citation of Related Structures:
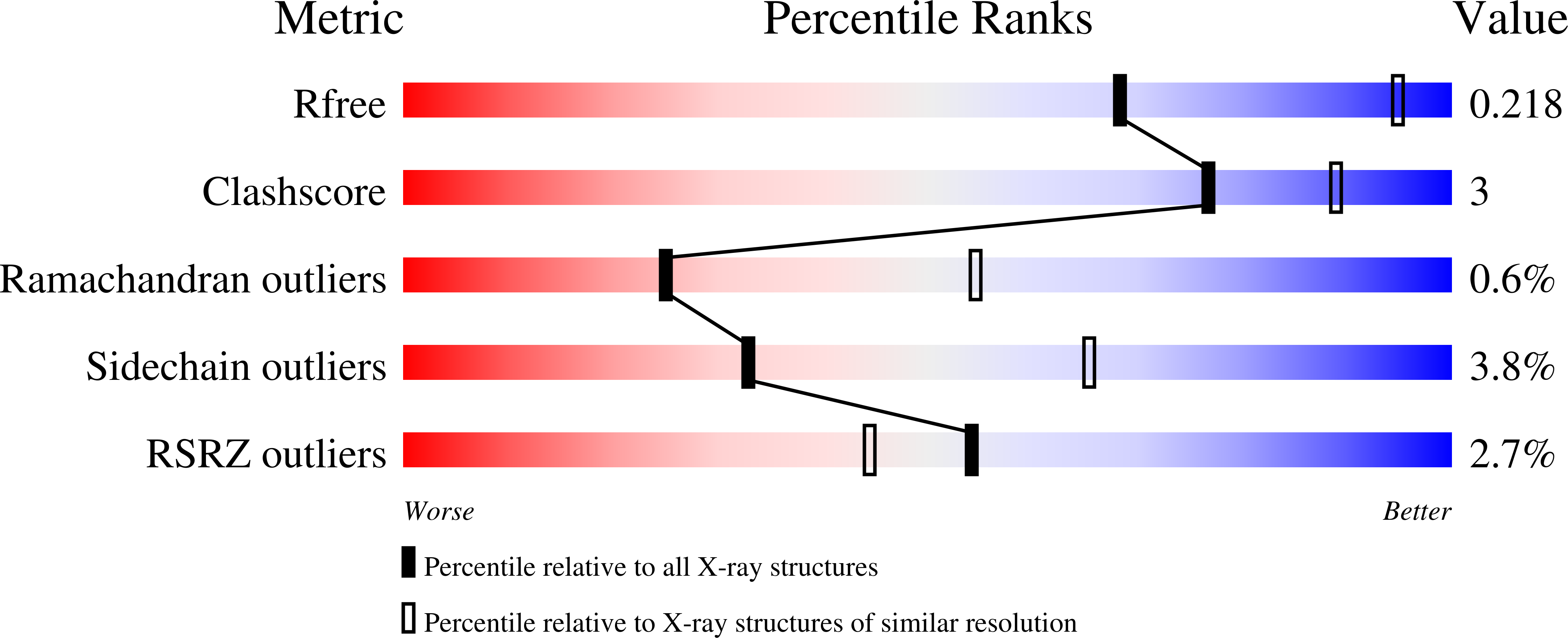
7NWK - PubMed Abstract:
Optimization of a series of azabenzimidazoles identified from screening hit 2 and the information gained from a co-crystal structure of the azabenzimidazole-based lead 6 bound to CDK9 led to the discovery of azaindoles as highly potent and selective CDK9 inhibitors. With the goal of discovering a highly selective and potent CDK9 inhibitor administrated intravenously that would enable transient target engagement of CDK9 for the treatment of hematological malignancies, further optimization focusing on physicochemical and pharmacokinetic properties led to azaindoles 38 and 39 . These compounds are highly potent and selective CDK9 inhibitors having short half-lives in rodents, suitable physical properties for intravenous administration, and the potential to achieve profound but transient inhibition of CDK9 in vivo .
Organizational Affiliation:
Oncology R&D, AstraZeneca, Cambridge CB4 0WG, United Kingdom.