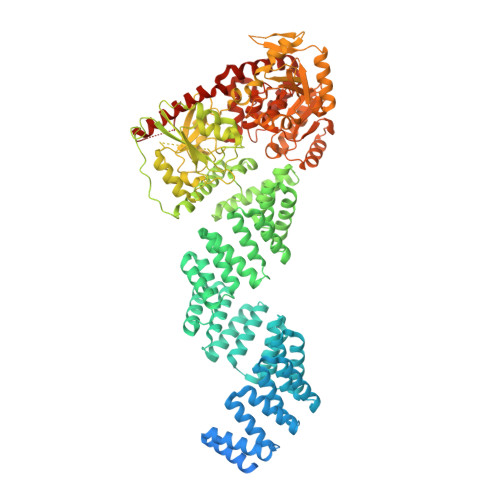
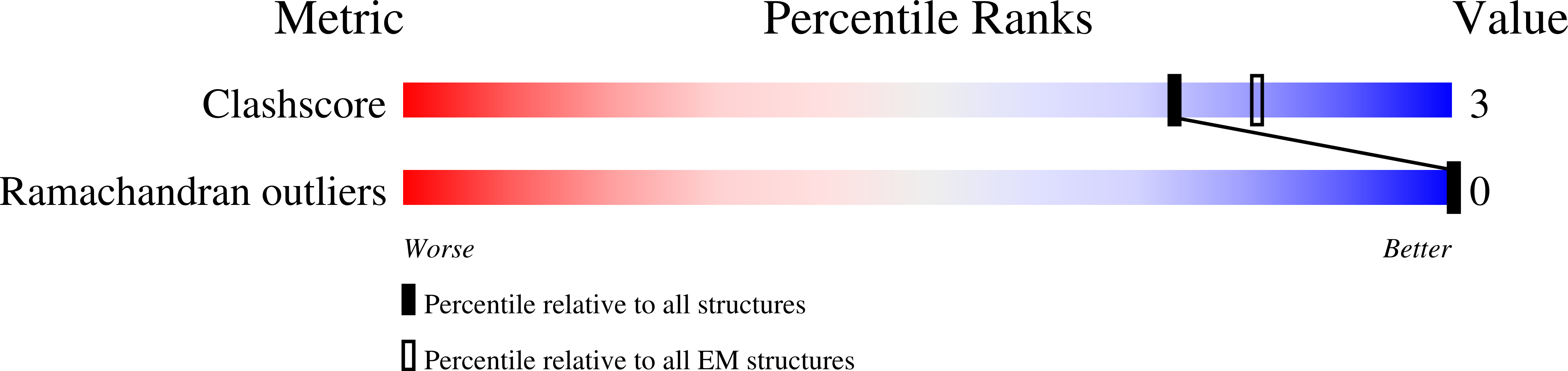Cryo-EM structure provides insights into the dimer arrangement of the O-linked beta-N-acetylglucosamine transferase OGT.
Meek, R.W., Blaza, J.N., Busmann, J.A., Alteen, M.G., Vocadlo, D.J., Davies, G.J.(2021) Nat Commun 12: 6508-6508
- PubMed: 34764280
- DOI: https://doi.org/10.1038/s41467-021-26796-6
- Primary Citation of Related Structures:
7NTF - PubMed Abstract:
The O-linked β-N-acetylglucosamine modification is a core signalling mechanism, with erroneous patterns leading to cancer and neurodegeneration. Although thousands of proteins are subject to this modification, only a single essential glycosyltransferase catalyses its installation, the O-GlcNAc transferase, OGT. Previous studies have provided truncated structures of OGT through X-ray crystallography, but the full-length protein has never been observed. Here, we report a 5.3 Å cryo-EM model of OGT. We show OGT is a dimer, providing a structural basis for how some X-linked intellectual disability mutations at the interface may contribute to disease. We observe that the catalytic section of OGT abuts a 13.5 tetratricopeptide repeat unit region and find the relative positioning of these sections deviate from the previously proposed, X-ray crystallography-based model. We also note that OGT exhibits considerable heterogeneity in tetratricopeptide repeat units N-terminal to the dimer interface with repercussions for how OGT binds protein ligands and partners.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, York, YO10 5DD, UK.