Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control.
Cottee, M.A., Beckwith, S.L., Letham, S.C., Kim, S.J., Young, G.R., Stoye, J.P., Garfinkel, D.J., Taylor, I.A.(2021) Nat Commun 12: 5590-5590
- PubMed: 34552077
- DOI: https://doi.org/10.1038/s41467-021-25849-0
- Primary Citation of Related Structures:
7NLG, 7NLH, 7NLI - PubMed Abstract:
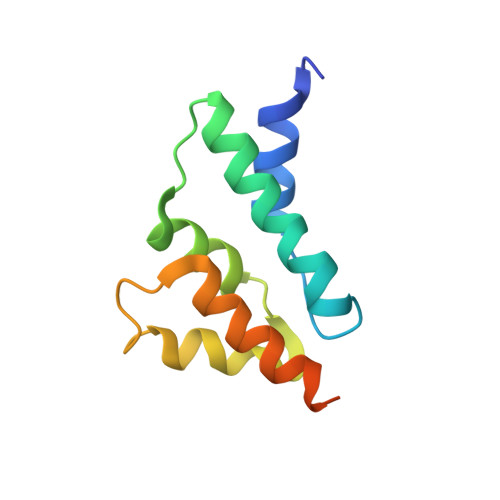
Excessive replication of Saccharomyces cerevisiae Ty1 retrotransposons is regulated by Copy Number Control, a process requiring the p22/p18 protein produced from a sub-genomic transcript initiated within Ty1 GAG. In retrotransposition, Gag performs the capsid functions required for replication and re-integration. To minimize genomic damage, p22/p18 interrupts virus-like particle function by interaction with Gag. Here, we present structural, biophysical and genetic analyses of p18m, a minimal fragment of Gag that restricts transposition. The 2.8 Å crystal structure of p18m reveals an all α-helical protein related to mammalian and insect ARC proteins. p18m retains the capacity to dimerise in solution and the crystal structures reveal two exclusive dimer interfaces. We probe our findings through biophysical analysis of interface mutants as well as Ty1 transposition and p18m restriction in vivo. Our data provide insight into Ty1 Gag structure and suggest how p22/p18 might function in restriction through a blocking-of-assembly mechanism.
Organizational Affiliation:
Macromolecular Structure Laboratory, The Francis Crick Institute, London, UK.