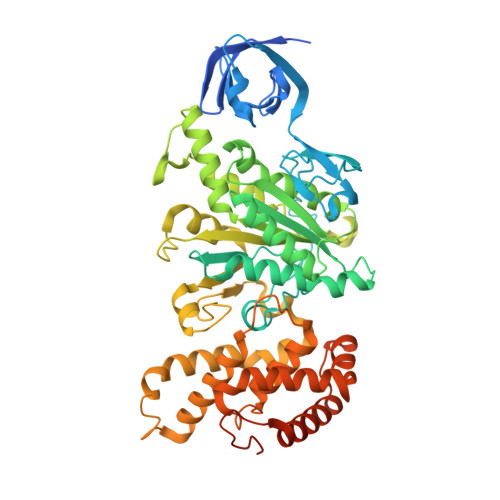
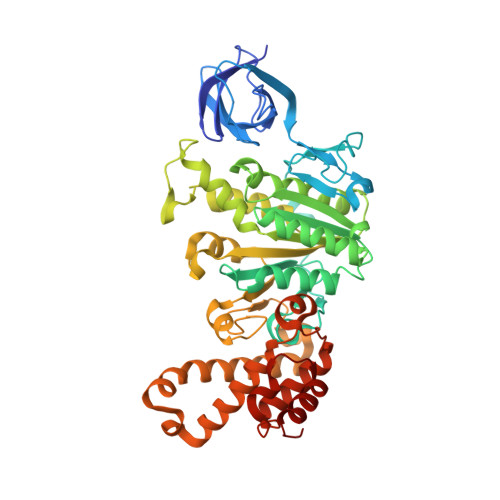
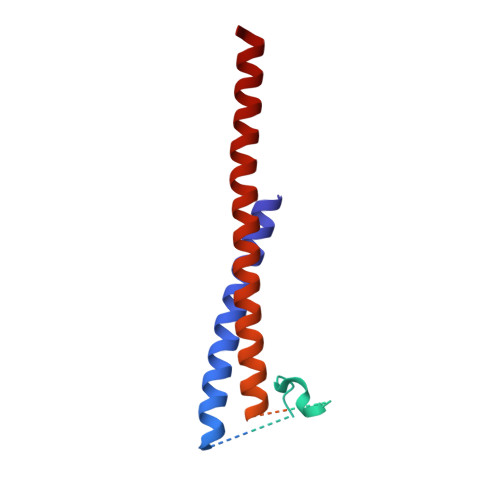
Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Montgomery, M.G., Petri, J., Spikes, T.E., Walker, J.E.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34782468
- DOI: https://doi.org/10.1073/pnas.2111899118
- Primary Citation of Related Structures:
7NJK, 7NJL, 7NJM, 7NJN, 7NJO, 7NJP, 7NJQ, 7NJR, 7NJS, 7NJT, 7NJU, 7NJV, 7NJW, 7NJX, 7NJY, 7NK7, 7NK9, 7NKB, 7NKD, 7NKH, 7NKJ, 7NKK, 7NKL, 7NKN, 7NKP, 7NKQ, 7NL9 - PubMed Abstract:
The structure has been determined by electron cryomicroscopy of the adenosine triphosphate (ATP) synthase from Mycobacterium smegmatis This analysis confirms features in a prior description of the structure of the enzyme, but it also describes other highly significant attributes not recognized before that are crucial for understanding the mechanism and regulation of the mycobacterial enzyme. First, we resolved not only the three main states in the catalytic cycle described before but also eight substates that portray structural and mechanistic changes occurring during a 360° catalytic cycle. Second, a mechanism of auto-inhibition of ATP hydrolysis involves not only the engagement of the C-terminal region of an α-subunit in a loop in the γ-subunit, as proposed before, but also a "fail-safe" mechanism involving the b'-subunit in the peripheral stalk that enhances engagement. A third unreported characteristic is that the fused bδ-subunit contains a duplicated domain in its N-terminal region where the two copies of the domain participate in similar modes of attachment of the two of three N-terminal regions of the α-subunits. The auto-inhibitory plus the associated "fail-safe" mechanisms and the modes of attachment of the α-subunits provide targets for development of innovative antitubercular drugs. The structure also provides support for an observation made in the bovine ATP synthase that the transmembrane proton-motive force that provides the energy to drive the rotary mechanism is delivered directly and tangentially to the rotor via a Grotthuss water chain in a polar L-shaped tunnel.
Organizational Affiliation:
The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Cambridge CB2 0XY, United Kingdom.