Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
Chaurasia, P., Nguyen, T.H.O., Rowntree, L.C., Juno, J.A., Wheatley, A.K., Kent, S.J., Kedzierska, K., Rossjohn, J., Petersen, J.(2021) J Biol Chem 297: 101065-101065
- PubMed: 34384783
- DOI: https://doi.org/10.1016/j.jbc.2021.101065
- Primary Citation of Related Structures:
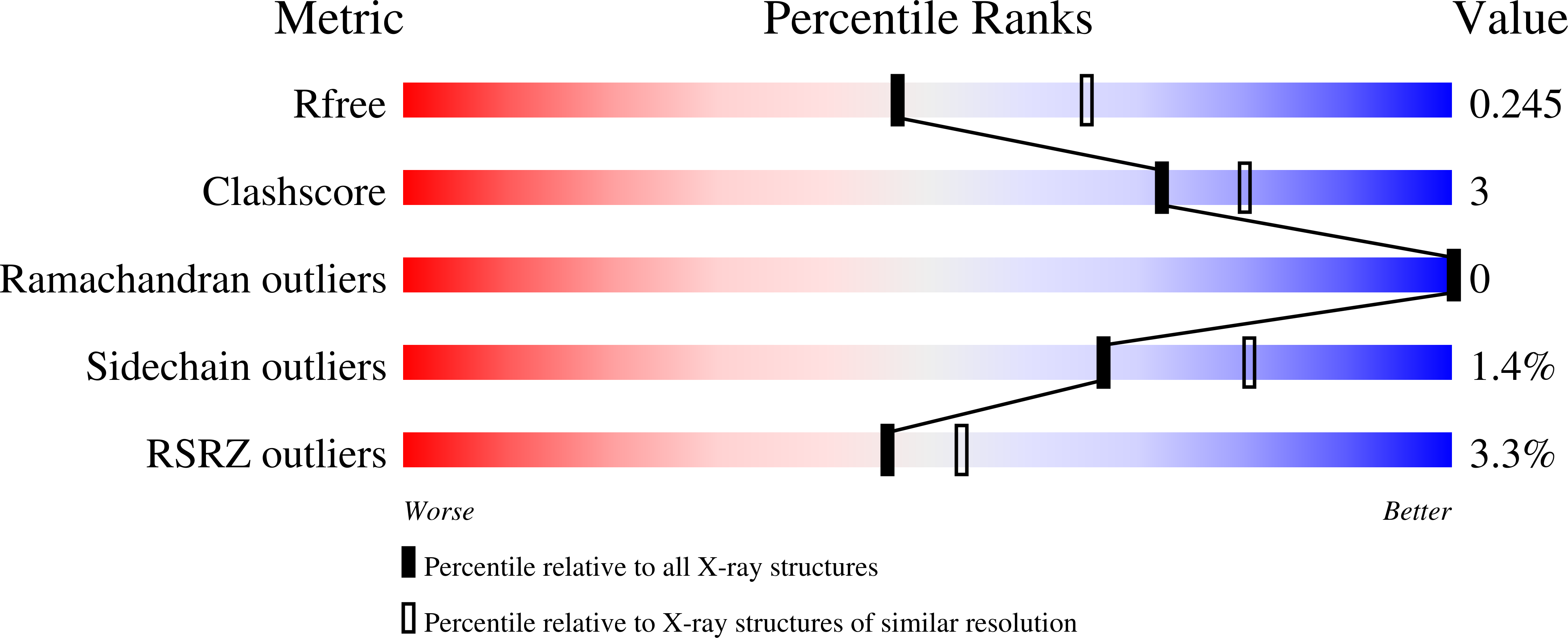
7N6D, 7N6E - PubMed Abstract:
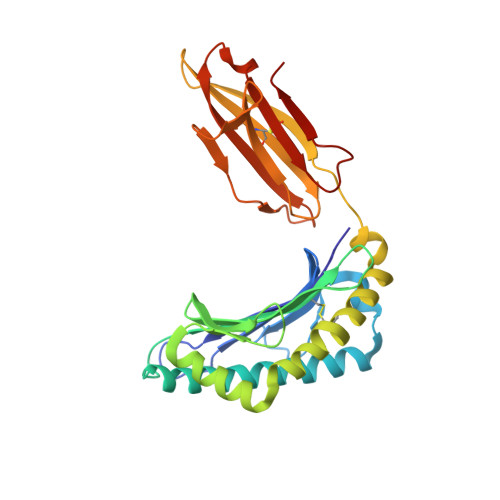
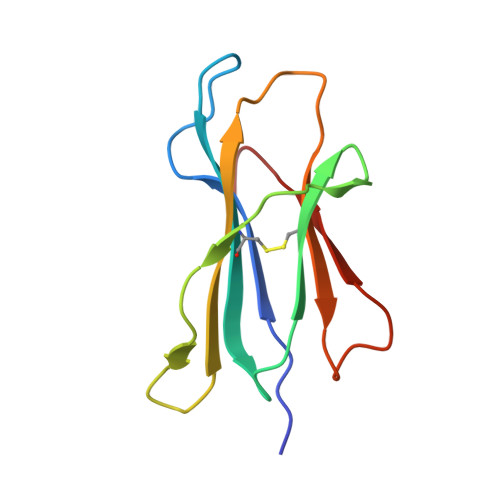
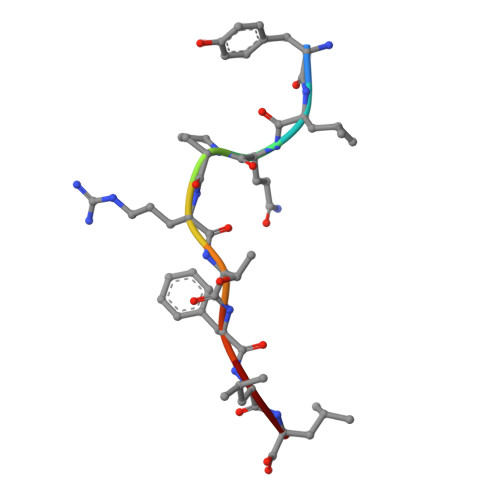
CD8 + T cells play an important role in vaccination and immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Although numerous SARS-CoV-2 CD8 + T cell epitopes have been identified, the molecular basis underpinning T cell receptor (TCR) recognition of SARS-CoV-2-specific T cells remains unknown. The T cell response directed toward SARS-CoV-2 spike protein-derived S 269-277 peptide presented by the human leukocyte antigen (HLA)-A∗02:01 allomorph (hereafter the HLA-A2 S269-277 epitope) is, to date, the most immunodominant SARS-CoV-2 epitope found in individuals bearing this allele. As HLA-A2 S269-277 -specific CD8 + T cells utilize biased TRAV12 gene usage within the TCR α-chain, we sought to understand the molecular basis underpinning this TRAV12 dominance. We expressed four TRAV12 + TCRs which bound the HLA-A2 S269-277 complex with low micromolar affinity and determined the crystal structure of the HLA-A2 S269-277 binary complex, and subsequently a ternary structure of the TRAV12 + TCR complexed to HLA-A2 S269-277 . We found that the TCR made extensive contacts along the entire length of the S 269-277 peptide, suggesting that the TRAV12 + TCRs would be sensitive to sequence variation within this epitope. To examine this, we investigated cross-reactivity toward analogous peptides from existing SARS-CoV-2 variants and closely related coronaviruses. We show via surface plasmon resonance and tetramer studies that the TRAV12 + T cell repertoire cross-reacts poorly with these analogous epitopes. Overall, we defined the structural basis underpinning biased TCR recognition of CD8 + T cells directed at an immunodominant epitope and provide a framework for understanding TCR cross-reactivity toward viral variants within the S 269-277 peptide.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.