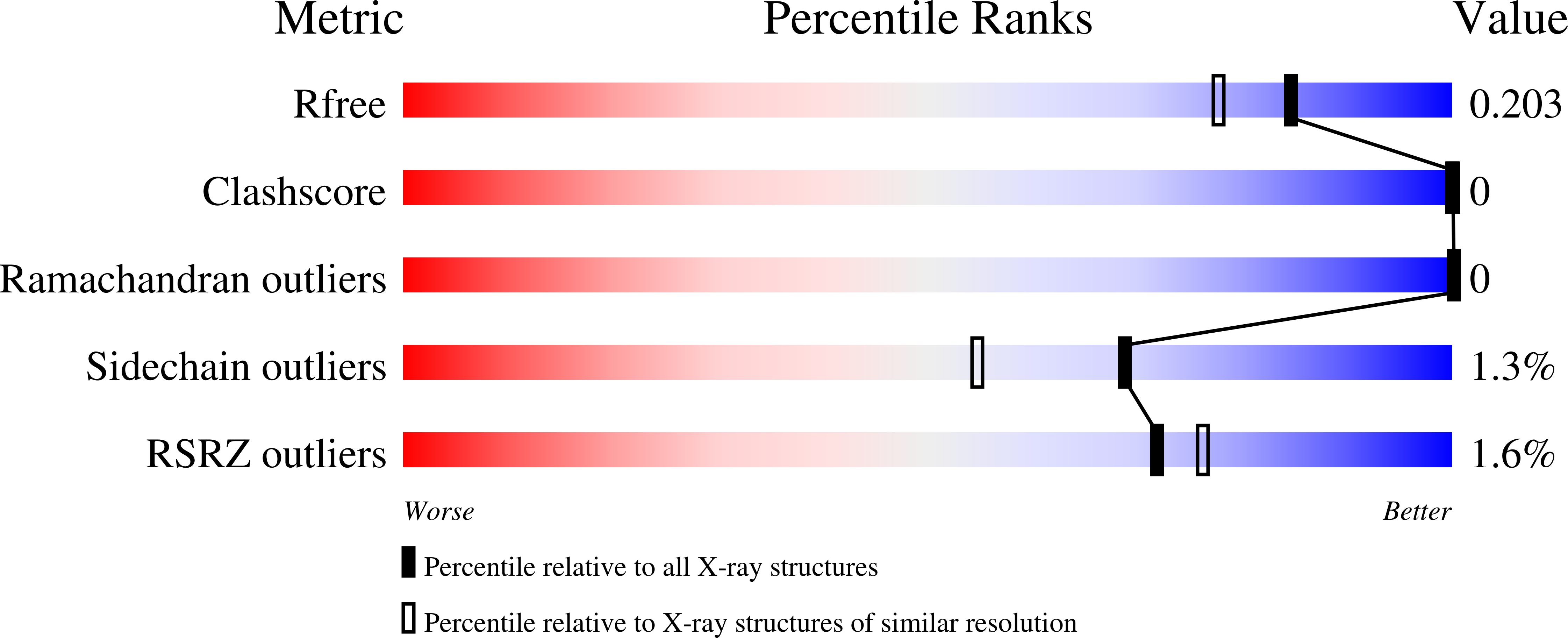
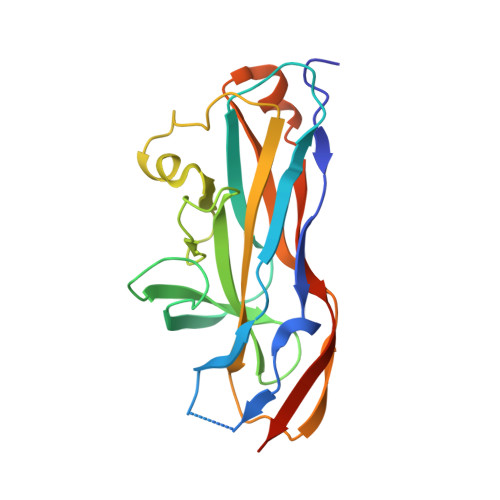
Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Hancock, S.J., Lo, A.W., Ve, T., Day, C.J., Tan, L., Mendez, A.A., Phan, M.D., Nhu, N.T.K., Peters, K.M., Richards, A.C., Fleming, B.A., Chang, C., Ngu, D.H.Y., Forde, B.M., Haselhorst, T., Goh, K.G.K., Beatson, S.A., Jennings, M.P., Mulvey, M.A., Kobe, B., Schembri, M.A.(2022) PLoS Pathog 18: e1010582-e1010582
- PubMed: 35700218
- DOI: https://doi.org/10.1371/journal.ppat.1010582
- Primary Citation of Related Structures:
7MZO, 7MZP, 7MZQ, 7MZR, 7MZS - PubMed Abstract:
Extra-intestinal pathogenic Escherichia coli (ExPEC) belong to a critical priority group of antibiotic resistant pathogens. ExPEC establish gut reservoirs that seed infection of the urinary tract and bloodstream, but the mechanisms of gut colonisation remain to be properly understood. Ucl fimbriae are attachment organelles that facilitate ExPEC adherence. Here, we investigated cellular receptors for Ucl fimbriae and Ucl expression to define molecular mechanisms of Ucl-mediated ExPEC colonisation of the gut. We demonstrate differential expression of Ucl fimbriae in ExPEC sequence types associated with disseminated infection. Genome editing of strains from two common sequence types, F11 (ST127) and UTI89 (ST95), identified a single nucleotide polymorphism in the ucl promoter that changes fimbriae expression via activation by the global stress-response regulator OxyR, leading to altered gut colonisation. Structure-function analysis of the Ucl fimbriae tip-adhesin (UclD) identified high-affinity glycan receptor targets, with highest affinity for sialyllacto-N-fucopentose VI, a structure likely to be expressed on the gut epithelium. Comparison of the UclD adhesin to the homologous UcaD tip-adhesin from Proteus mirabilis revealed that although they possess a similar tertiary structure, apart from lacto-N-fucopentose VI that bound to both adhesins at low-micromolar affinity, they recognize different fucose- and glucose-containing oligosaccharides. Competitive surface plasmon resonance analysis together with co-structural investigation of UcaD in complex with monosaccharides revealed a broad-specificity glycan binding pocket shared between UcaD and UclD that could accommodate these interactions. Overall, our study describes a mechanism of adaptation that augments establishment of an ExPEC gut reservoir to seed disseminated infections, providing a pathway for the development of targeted anti-adhesion therapeutics.
Organizational Affiliation:
School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Queensland, Australia.