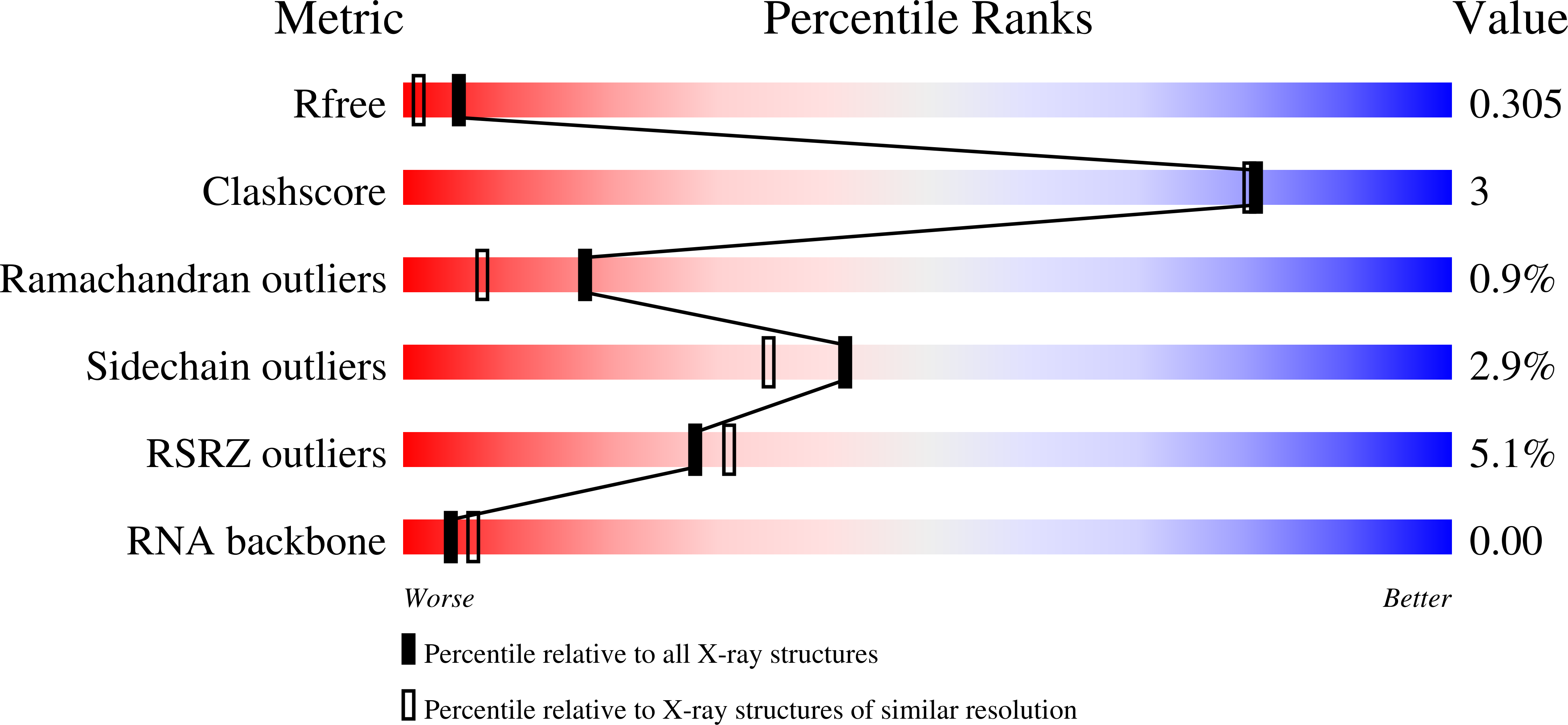
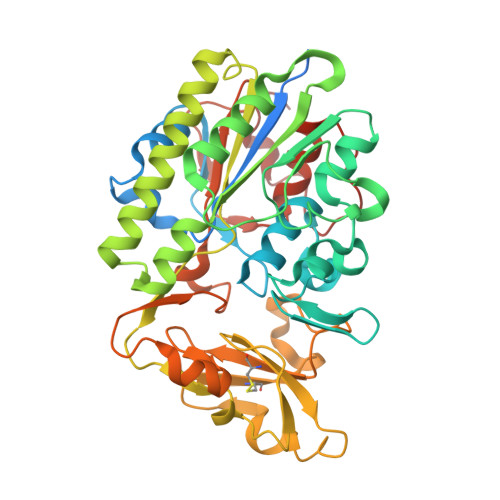
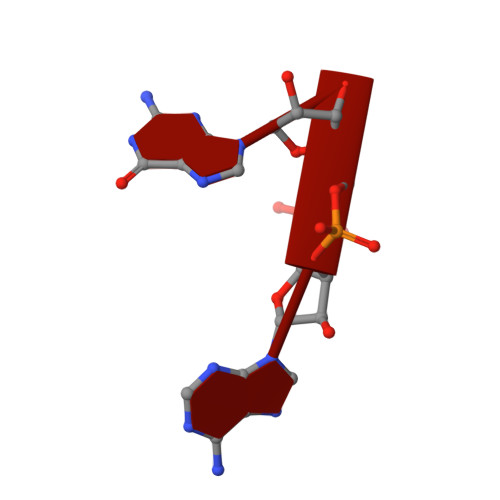
ENPP1's regulation of extracellular cGAMP is a ubiquitous mechanism of attenuating STING signaling.
Carozza, J.A., Cordova, A.F., Brown, J.A., AlSaif, Y., Bohnert, V., Cao, X., Mardjuki, R.E., Skariah, G., Fernandez, D., Li, L.(2022) Proc Natl Acad Sci U S A 119: e2119189119-e2119189119
- PubMed: 35588451
- DOI: https://doi.org/10.1073/pnas.2119189119
- Primary Citation of Related Structures:
7MW8, 7N1S - PubMed Abstract:
The metazoan innate immune second messenger 2′3′-cGAMP is present both inside and outside cells. However, only extracellular cGAMP can be negatively regulated by the extracellular hydrolase ENPP1. Here, we determine whether ENPP1’s regulation of extracellular cGAMP is a ubiquitous mechanism of attenuating stimulator of interferon genes (STING) signaling. We identified ENPP1H362A, a point mutation that cannot degrade the 2′-5′ linkage in cGAMP while maintaining otherwise normal function. The selectivity of this histidine is conserved down to bacterial nucleotide pyrophosphatase/phosphodiesterase (NPP), allowing structural analysis and suggesting an unexplored ancient history of 2′-5′ cyclic dinucleotides. Enpp1H362A mice demonstrated that extracellular cGAMP is not responsible for the devastating phenotype in ENPP1-null humans and mice but is responsible for antiviral immunity and systemic inflammation. Our data define extracellular cGAMP as a pivotal STING activator, identify an evolutionarily critical role for ENPP1 in regulating inflammation, and suggest a therapeutic strategy for viral and inflammatory conditions by manipulating ENPP1 activity.
Organizational Affiliation:
ChEM-H Institute, Stanford University, Stanford, CA 94301.