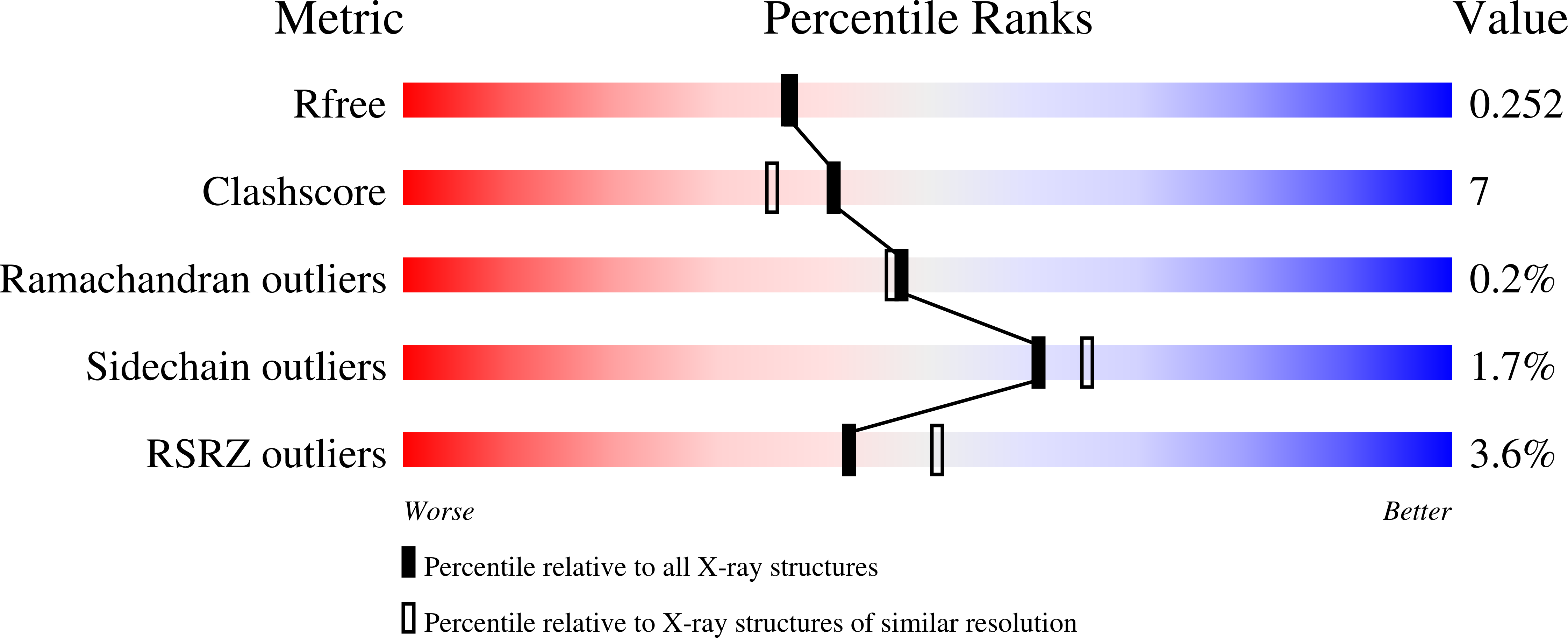
Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7.
Sevrioukova, I.F.(2021) Int J Mol Sci 22
- PubMed: 34072457
- DOI: https://doi.org/10.3390/ijms22115831
- Primary Citation of Related Structures:
7MK8 - PubMed Abstract:
Cytochrome P450 3A7 (CYP3A7) is a fetal/neonatal liver enzyme that participates in estriol synthesis, clearance of all-trans retinoic acid, and xenobiotic metabolism. Compared to the closely related major drug-metabolizing enzyme in adult liver, CYP3A4, the ligand binding and catalytic capacity of CYP3A7 are substantially reduced. To better understand the structural basis for these functional differences, the 2.15 Å crystal structure of CYP3A7 has been solved. Comparative analysis of CYP3A enzymes shows that decreased structural plasticity rather than the active site microenvironment defines the ligand binding ability of CYP3A7. In particular, a rotameric switch in the gatekeeping amino acid F304 triggers local and long-range rearrangements that transmit to the F-G fragment and alter its interactions with the I-E-D-helical core, resulting in a more rigid structure. Elongation of the β 3 -β 4 strands, H-bond linkage in the substrate channel, and steric constraints in the C-terminal loop further increase the active site rigidity and limit conformational ensemble. Collectively, these structural distinctions lower protein plasticity and change the heme environment, which, in turn, could impede the spin-state transition essential for optimal reactivity and oxidation of substrates.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900, USA.