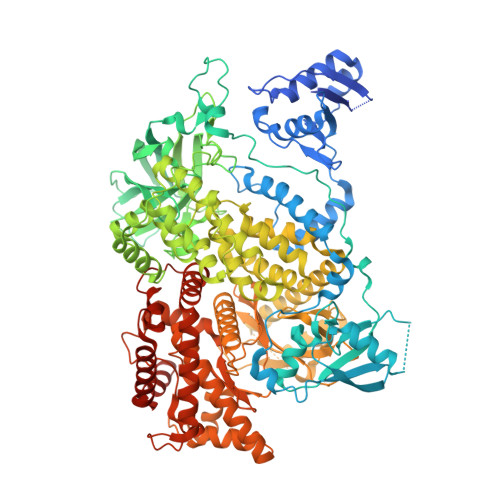
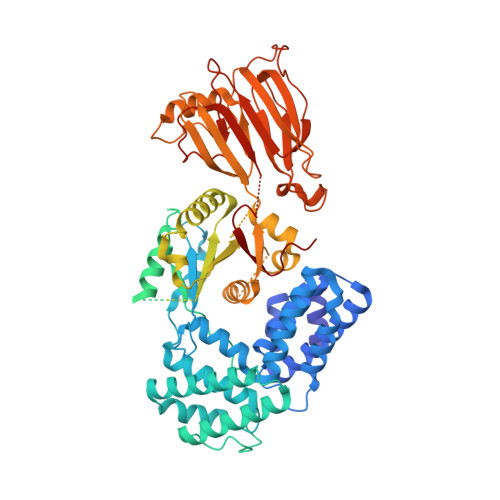
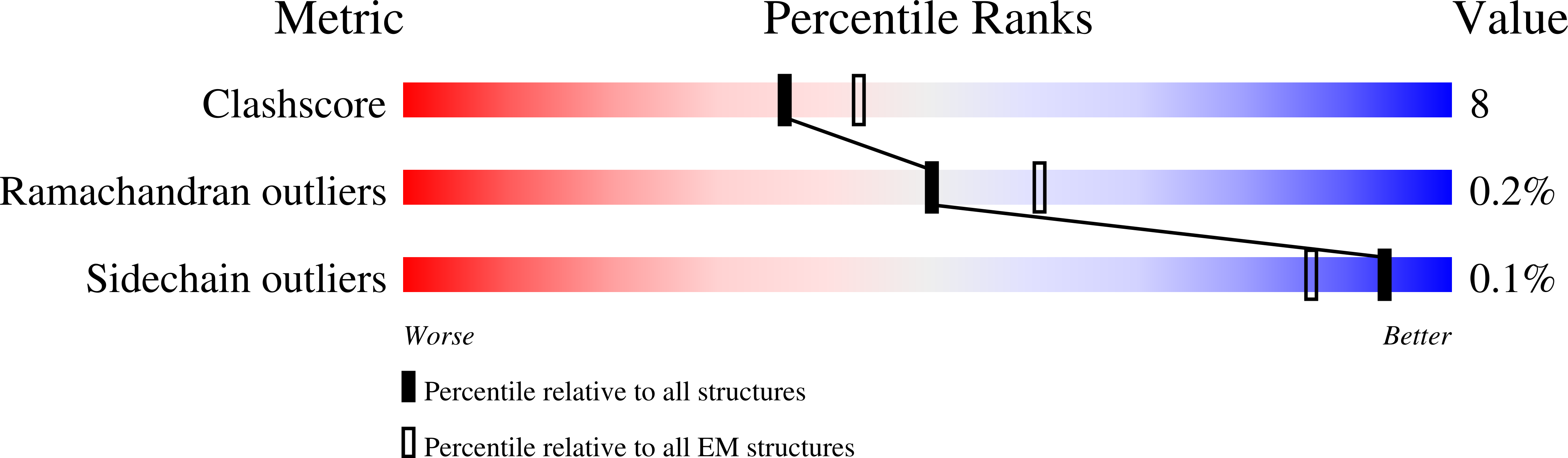Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Rathinaswamy, M.K., Dalwadi, U., Fleming, K.D., Adams, C., Stariha, J.T.B., Pardon, E., Baek, M., Vadas, O., DiMaio, F., Steyaert, J., Hansen, S.D., Yip, C.K., Burke, J.E.(2021) Sci Adv 7
- PubMed: 34452907
- DOI: https://doi.org/10.1126/sciadv.abj4282
- Primary Citation of Related Structures:
7MEZ - PubMed Abstract:
The class IB phosphoinositide 3-kinase (PI3K), PI3Kγ, is a master regulator of immune cell function and a promising drug target for both cancer and inflammatory diseases. Critical to PI3Kγ function is the association of the p110γ catalytic subunit to either a p101 or p84 regulatory subunit, which mediates activation by G protein-coupled receptors. Here, we report the cryo-electron microscopy structure of a heterodimeric PI3Kγ complex, p110γ-p101. This structure reveals a unique assembly of catalytic and regulatory subunits that is distinct from other class I PI3K complexes. p101 mediates activation through its Gβγ-binding domain, recruiting the heterodimer to the membrane and allowing for engagement of a secondary Gβγ-binding site in p110γ. Mutations at the p110γ-p101 and p110γ-adaptor binding domain interfaces enhanced Gβγ activation. A nanobody that specifically binds to the p101-Gβγ interface blocks activation, providing a novel tool to study and target p110γ-p101-specific signaling events in vivo.
Organizational Affiliation:
Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia, Canada.