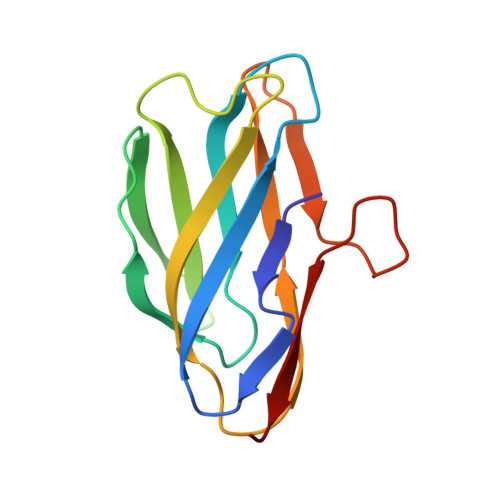
The YcnI protein from Bacillus subtilis contains a copper-binding domain.
Damle, M.S., Singh, A.N., Peters, S.C., Szalai, V.A., Fisher, O.S.(2021) J Biol Chem 297: 101078-101078
- PubMed: 34400169
- DOI: https://doi.org/10.1016/j.jbc.2021.101078
- Primary Citation of Related Structures:
7ME6, 7MEK - PubMed Abstract:
Bacteria require a precise balance of copper ions to ensure that essential cuproproteins are fully metalated while also avoiding copper-induced toxicity. The Gram-positive bacterium Bacillus subtilis maintains appropriate copper homeostasis in part through the ycn operon. The ycn operon comprises genes encoding three proteins: the putative copper importer YcnJ, the copper-dependent transcriptional repressor YcnK, and the uncharacterized Domain of Unknown Function 1775 (DUF1775) containing YcnI. DUF1775 domains are found across bacterial phylogeny, and bioinformatics analyses indicate that they frequently neighbor domains implicated in copper homeostasis and transport. Here, we investigated whether YcnI can interact with copper and, using electron paramagnetic resonance and inductively coupled plasma-MS, found that this protein can bind a single Cu(II) ion. We determine the structure of both the apo and copper-bound forms of the protein by X-ray crystallography, uncovering a copper-binding site featuring a unique monohistidine brace ligand set that is highly conserved among DUF1775 domains. These data suggest a possible role for YcnI as a copper chaperone and that DUF1775 domains in other bacterial species may also function in copper homeostasis.
Organizational Affiliation:
Department of Chemistry, Lehigh University, Bethlehem, Pennsylvania, USA.