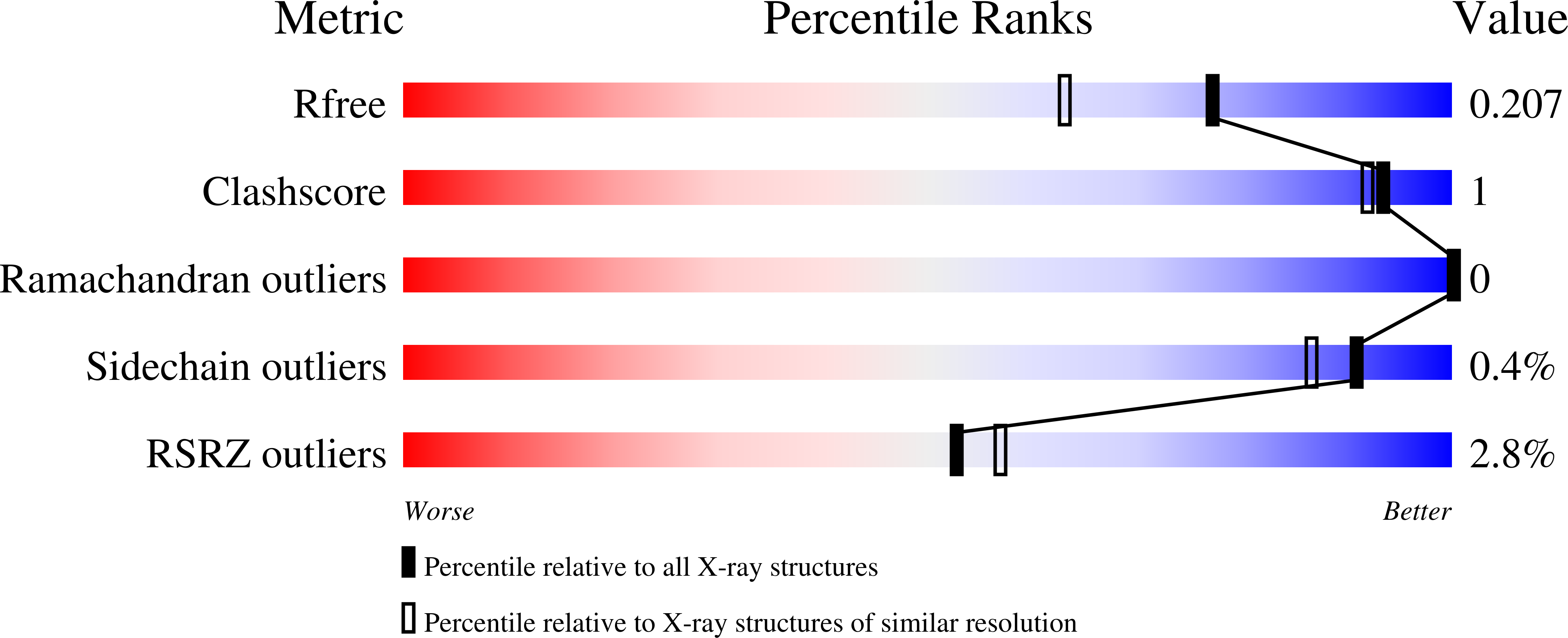
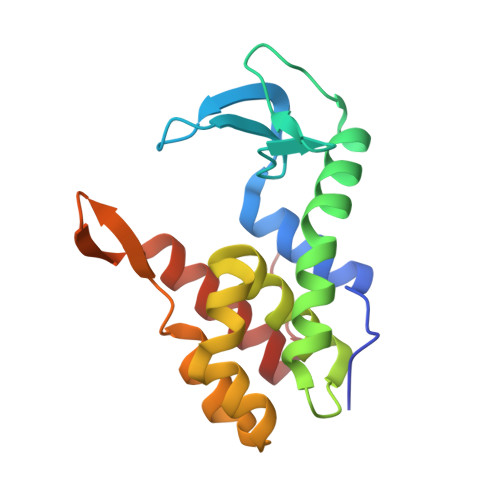
The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Love, M.J., Coombes, D., Manners, S.H., Abeysekera, G.S., Billington, C., Dobson, R.C.J.(2021) Viruses 13
- PubMed: 34207694
- DOI: https://doi.org/10.3390/v13061101
- Primary Citation of Related Structures:
7M5I - PubMed Abstract:
Bacteriophage-encoded endolysins have been identified as antibacterial candidates. However, the development of endolysins as mainstream antibacterial agents first requires a comprehensive biochemical understanding. This study defines the atomic structure and enzymatic function of Escherichia coli O157:H7 phage FAHEc1 endolysin, LysF1. Bioinformatic analysis suggests this endolysin belongs to the T4 Lysozyme (T4L)-like family of proteins and contains a highly conserved catalytic triad. We then solved the structure of LysF1 with x-ray crystallography to 1.71 Å. LysF1 was confirmed to exist as a monomer in solution by sedimentation velocity experiments. The protein architecture of LysF1 is conserved between T4L and related endolysins. Comparative analysis with related endolysins shows that the spatial orientation of the catalytic triad is conserved, suggesting the catalytic mechanism of peptidoglycan degradation is the same as that of T4L. Differences in the sequence illustrate the role coevolution may have in the evolution of this fold. We also demonstrate that by mutating a single residue within the hydrophobic core, the thermal stability of LysF1 can be increased by 9.4 °C without compromising enzymatic activity. Overall, the characterization of LysF1 provides further insight into the T4L-like class of endolysins. Our study will help advance the development of related endolysins as antibacterial agents, as rational engineering will rely on understanding mutable positions within this protein fold.
Organizational Affiliation:
Biomolecular Interaction Centre and School of Biological Sciences, University of Canterbury, Christchurch 8041, New Zealand.