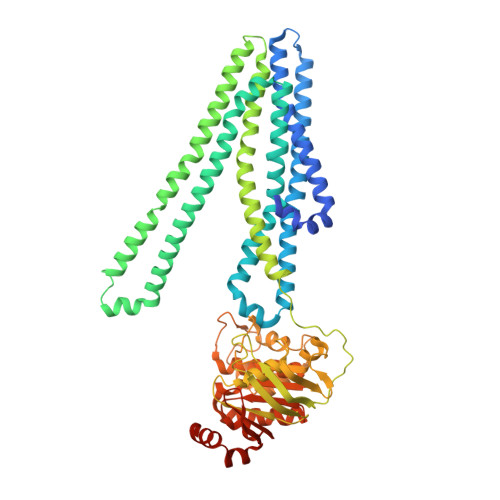
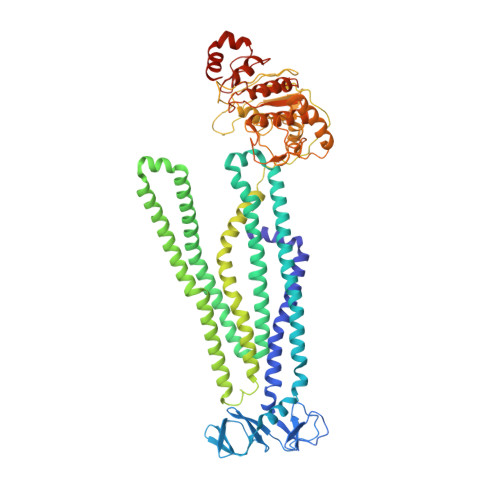
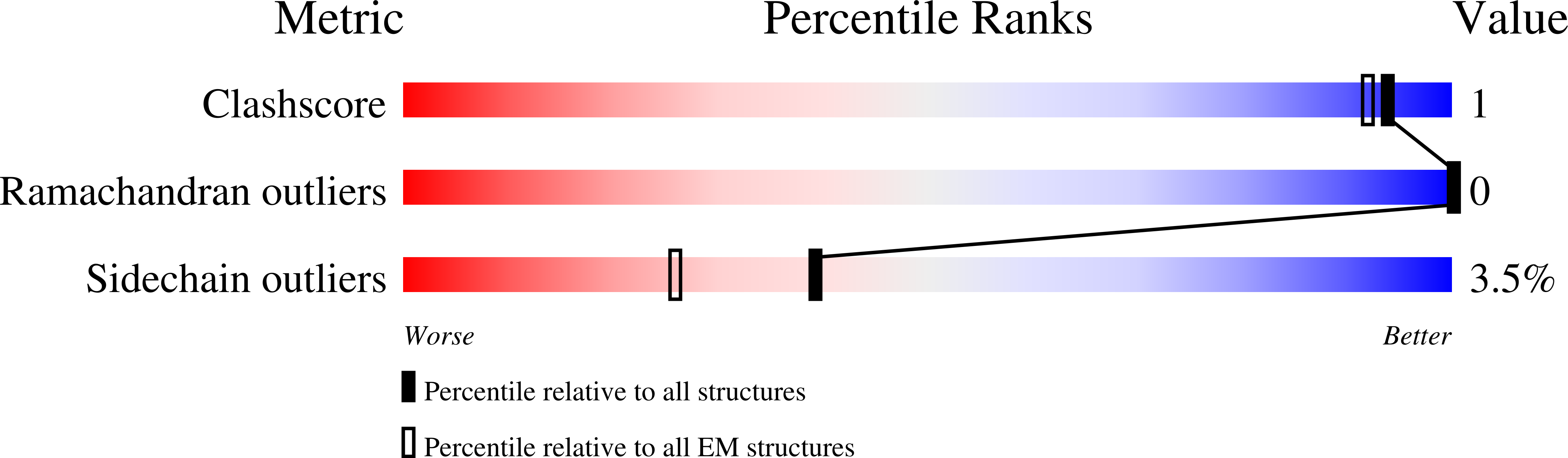Asymmetric drug binding in an ATP-loaded inward-facing state of an ABC transporter.
Thaker, T.M., Mishra, S., Zhou, W., Mohan, M., Tang, Q., Faraldo-Gomez, J.D., Mchaourab, H.S., Tomasiak, T.M.(2022) Nat Chem Biol 18: 226-235
- PubMed: 34931066
- DOI: https://doi.org/10.1038/s41589-021-00936-x
- Primary Citation of Related Structures:
7M33 - PubMed Abstract:
Substrate efflux by ATP-binding cassette (ABC) transporters, which play a major role in multidrug resistance, entails the ATP-powered interconversion between transporter intermediates. Despite recent progress in structure elucidation, a number of intermediates have yet to be visualized and mechanistically interpreted. Here, we combine cryogenic-electron microscopy (cryo-EM), double electron-electron resonance spectroscopy and molecular dynamics simulations to profile a previously unobserved intermediate of BmrCD, a heterodimeric multidrug ABC exporter from Bacillus subtilis. In our cryo-EM structure, ATP-bound BmrCD adopts an inward-facing architecture featuring two molecules of the substrate Hoechst-33342 in a striking asymmetric head-to-tail arrangement. Deletion of the extracellular domain capping the substrate-binding chamber or mutation of Hoechst-coordinating residues abrogates cooperative stimulation of ATP hydrolysis. Together, our findings support a mechanistic role for symmetry mismatch between the nucleotide binding and the transmembrane domains in the conformational cycle of ABC transporters and is of notable importance for rational design of molecules for targeted ABC transporter inhibition.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ, USA.