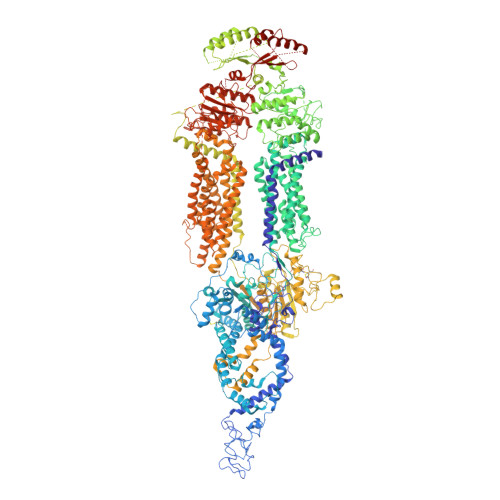
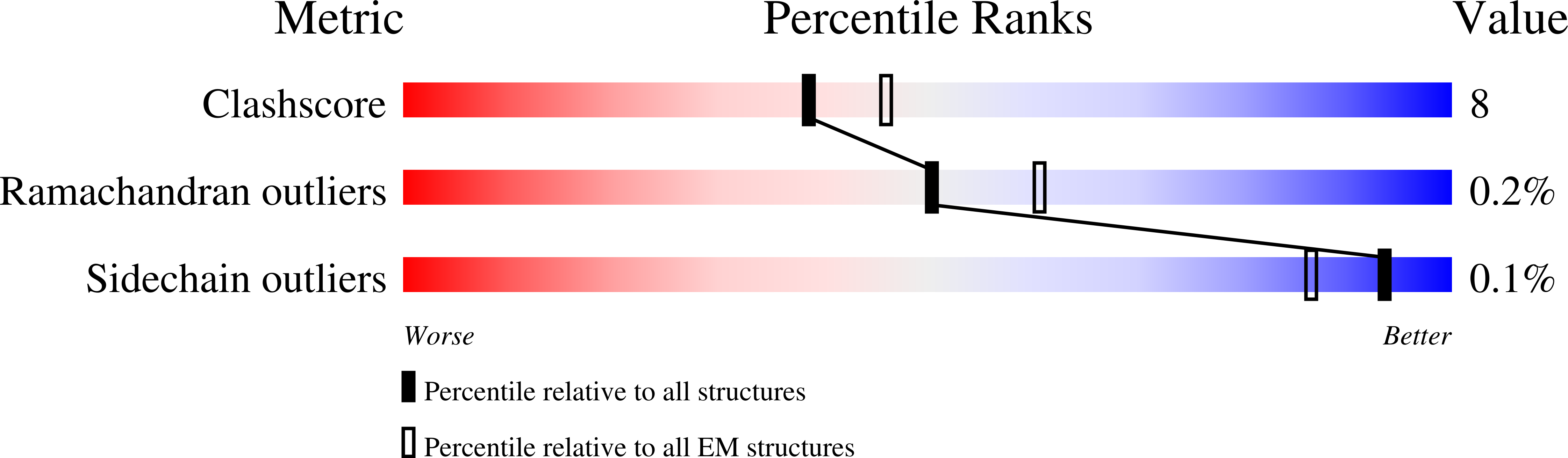Cryo-EM structures of the ABCA4 importer reveal mechanisms underlying substrate binding and Stargardt disease.
Scortecci, J.F., Molday, L.L., Curtis, S.B., Garces, F.A., Panwar, P., Van Petegem, F., Molday, R.S.(2021) Nat Commun 12: 5902-5902
- PubMed: 34625547
- DOI: https://doi.org/10.1038/s41467-021-26161-7
- Primary Citation of Related Structures:
7M1P, 7M1Q - PubMed Abstract:
ABCA4 is an ATP-binding cassette (ABC) transporter that flips N-retinylidene-phosphatidylethanolamine (N-Ret-PE) from the lumen to the cytoplasmic leaflet of photoreceptor membranes. Loss-of-function mutations cause Stargardt disease (STGD1), a macular dystrophy associated with severe vision loss. To define the mechanisms underlying substrate binding and STGD1, we determine the cryo-EM structure of ABCA4 in its substrate-free and bound states. The two structures are similar and delineate an elongated protein with the two transmembrane domains (TMD) forming an outward facing conformation, extended and twisted exocytoplasmic domains (ECD), and closely opposed nucleotide binding domains. N-Ret-PE is wedged between the two TMDs and a loop from ECD1 within the lumen leaflet consistent with a lateral access mechanism and is stabilized through hydrophobic and ionic interactions with residues from the TMDs and ECDs. Our studies provide a framework for further elucidating the molecular mechanism associated with lipid transport and disease and developing promising disease interventions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada.