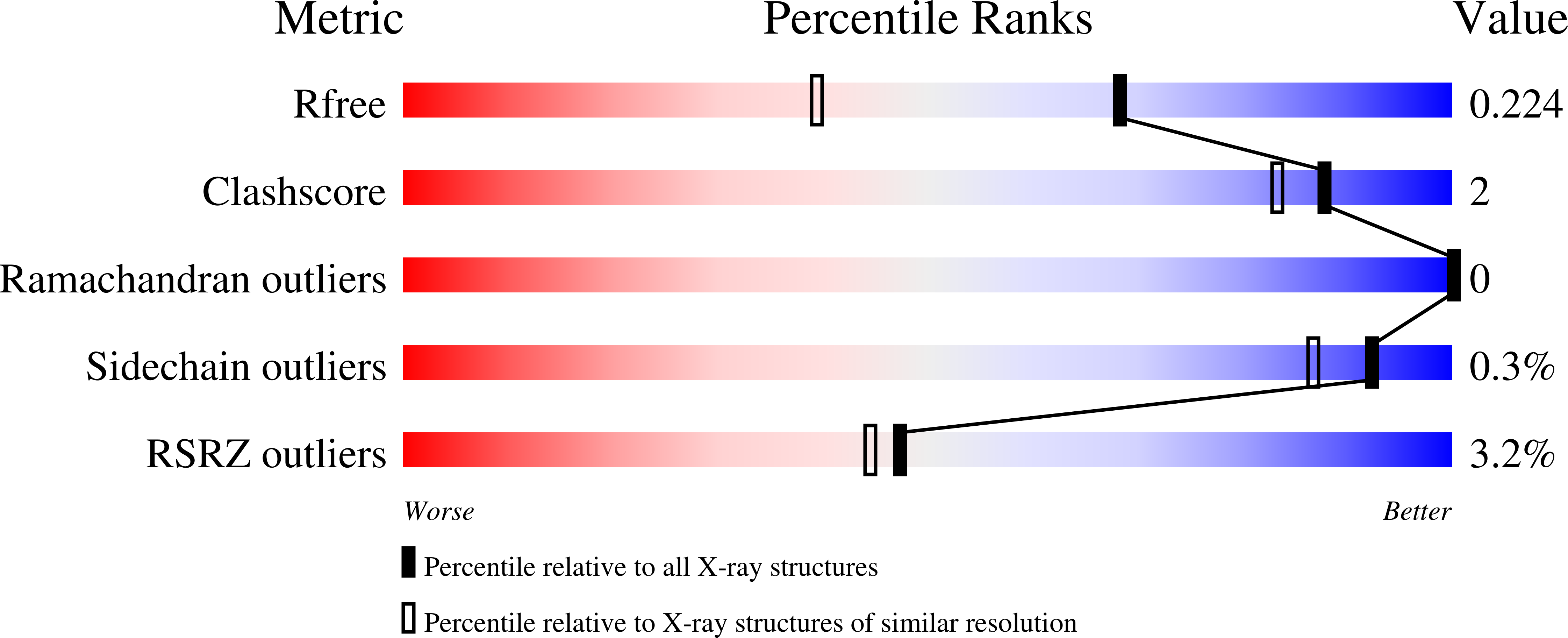
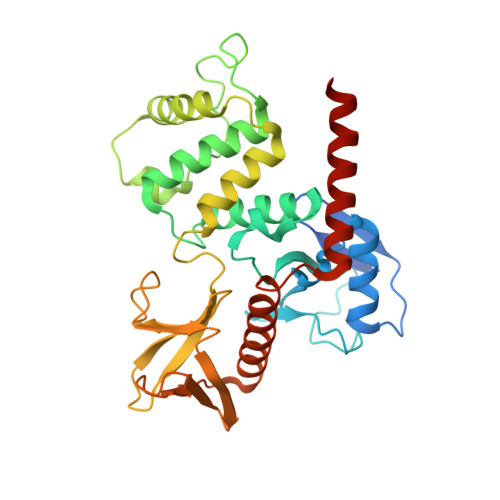
Conformational flexibility determines the Nf2/merlin tumor suppressor functions.
Primi, M.C., Rangarajan, E.S., Patil, D.N., Izard, T.(2021) Matrix Biol Plus 12: 100074-100074
- PubMed: 34337379
- DOI: https://doi.org/10.1016/j.mbplus.2021.100074
- Primary Citation of Related Structures:
7LWH - PubMed Abstract:
The Neurofibromatosis type 2 gene encodes the Nf2/merlin tumor suppressor protein that is responsible for the regulation of cell proliferation. Once activated, Nf2/merlin modulates adhesive signaling pathways and thereby inhibits cell growth. Nf2/merlin controls oncogenic gene expression by modulating the Hippo pathway. By responding to several physical and biochemical stimuli, Hippo signaling determines contact inhibition of proliferation as well as organ size. The large tumor suppressor (LATS) serine/threonine-protein kinase is the key enzyme in the highly conserved kinase cascade that negatively regulates the activity and localization of the transcriptional coactivators Yes-associated protein (YAP) and its paralogue transcriptional coactivator with PDZ-binding motif (TAZ). Nf2/merlin belongs to the band 4.1, ezrin, radixin, moesin (FERM) gene family that links the actin cytoskeleton to adherens junctions, remodels adherens junctions during epithelial morphogenesis and maintains organized apical surfaces on the plasma cell membrane. Nf2/merlin and ERM proteins have a globular N -terminal cloverleaf head domain, the FERM domain, that binds to the plasma membrane, a central α-helical domain, and a tail domain that binds to its head domain. Here we present the high-resolution crystal structure of Nf2/merlin bound to LATS1 which shows that LATS1 binding to Nf2/merlin displaces the Nf2/merlin tail domain and causes an allosteric shift in the Nf2/merlin α-helix that extends from its FERM domain. This is consistent with the fact that full-length Nf2/merlin binds LATS1 ~10-fold weaker compared to LATS1 binding to the Nf2/merlin-PIP 2 complex. Our data increase our understanding of Nf2/merlin biology by providing mechanistic insights into the Hippo pathway that are relevant to several diseases in particular oncogenic features that are associated with cancers.
Organizational Affiliation:
Cell Adhesion Laboratory, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, Jupiter 33458, FL, United States.