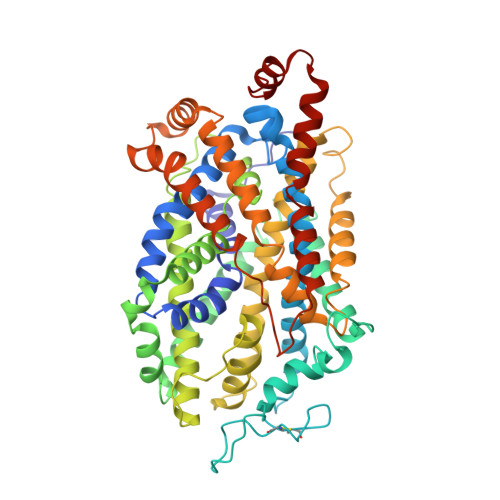
The antidepressant drug vilazodone is an allosteric inhibitor of the serotonin transporter.
Plenge, P., Yang, D., Salomon, K., Laursen, L., Kalenderoglou, I.E., Newman, A.H., Gouaux, E., Coleman, J.A., Loland, C.J.(2021) Nat Commun 12: 5063-5063
- PubMed: 34417466
- DOI: https://doi.org/10.1038/s41467-021-25363-3
- Primary Citation of Related Structures:
7LWD - PubMed Abstract:
Depression is a common mental disorder. The standard medical treatment is the selective serotonin reuptake inhibitors (SSRIs). All characterized SSRIs are competitive inhibitors of the serotonin transporter (SERT). A non-competitive inhibitor may produce a more favorable therapeutic profile. Vilazodone is an antidepressant with limited information on its molecular interactions with SERT. Here we use molecular pharmacology and cryo-EM structural elucidation to characterize vilazodone binding to SERT. We find that it exhibits non-competitive inhibition of serotonin uptake and impedes dissociation of [ 3 H]imipramine at low nanomolar concentrations. Our SERT structure with bound imipramine and vilazodone reveals a unique binding pocket for vilazodone, expanding the boundaries of the extracellular vestibule. Characterization of the binding site is substantiated with molecular dynamics simulations and systematic mutagenesis of interacting residues resulting in decreased vilazodone binding to the allosteric site. Our findings underline the versatility of SERT allosteric ligands and describe the unique binding characteristics of vilazodone.
Organizational Affiliation:
Laboratory for Membrane Protein Dynamics. Department of Neuroscience, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.