Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Feng, M., Harijan, R.K., Harris, L.D., Tyler, P.C., Frohlich, R.F.G., Brown, M., Schramm, V.L.(2021) Biochemistry 60: 1933-1946
- PubMed: 34077175
- DOI: https://doi.org/10.1021/acs.biochem.1c00215
- Primary Citation of Related Structures:
7LKJ, 7LKK - PubMed Abstract:
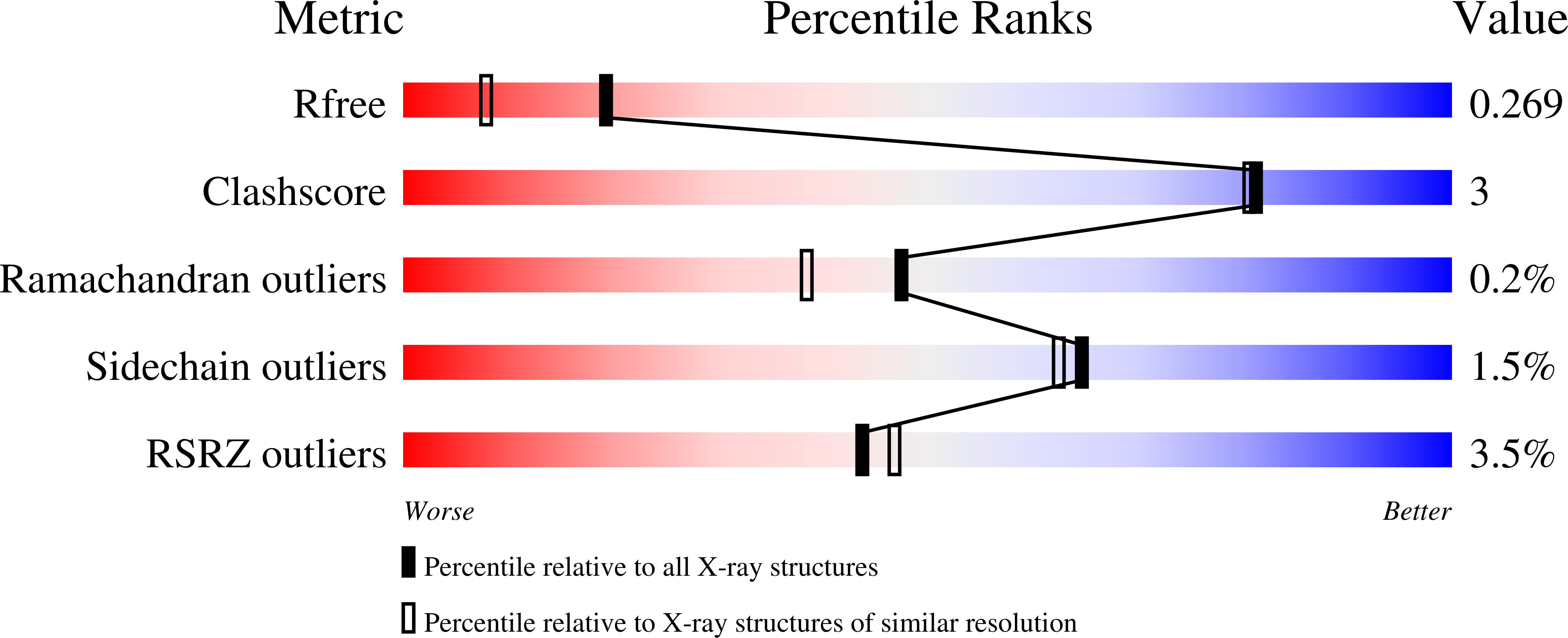
Helicobacter pylori is a Gram-negative bacterium that is responsible for gastric and duodenal ulcers. H. pylori uses the unusual mqn pathway with aminofutalosine (AFL) as an intermediate for menaquinone biosynthesis. Previous reports indicate that hydrolysis of AFL by 5'-methylthioadenosine nucleosidase ( Hp MTAN) is the direct path for producing downstream metabolites in the mqn pathway. However, genomic analysis indicates jhp0252 is a candidate for encoding AFL deaminase (AFLDA), an activity for deaminating aminofutolasine. The product, futalosine, is not a known substrate for bacterial MTANs. Recombinant jhp0252 was expressed and characterized as an AFL deaminase ( Hp AFLDA). Its catalytic specificity includes AFL, 5'-methylthioadenosine, 5'-deoxyadenosine, adenosine, and S -adenosylhomocysteine. The k cat / K m value for AFL is 6.8 × 10 4 M -1 s -1 , 26-fold greater than that for adenosine. 5'-Methylthiocoformycin (MTCF) is a slow-onset inhibitor for Hp AFLDA and demonstrated inhibitory effects on H. pylori growth. Supplementation with futalosine partially restored H. pylori growth under MTCF treatment, suggesting AFL deamination is significant for cell growth. The crystal structures of apo- Hp AFLDA and with MTCF at the catalytic sites show a catalytic site Zn 2+ or Fe 2+ as the water-activating group. With bound MTCF, the metal ion is 2.0 Å from the sp 3 hydroxyl group of the transition state analogue. Metabolomics analysis revealed that Hp AFLDA has intracellular activity and is inhibited by MTCF. The mqn pathway in H. pylori bifurcates at aminofutalosine with Hp MTAN producing adenine and depurinated futalosine and Hp AFLDA producing futalosine. Inhibition of cellular Hp MTAN or Hp AFLDA decreased the cellular content of menaquinone-6, supporting roles for both enzymes in the pathway.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, United States.