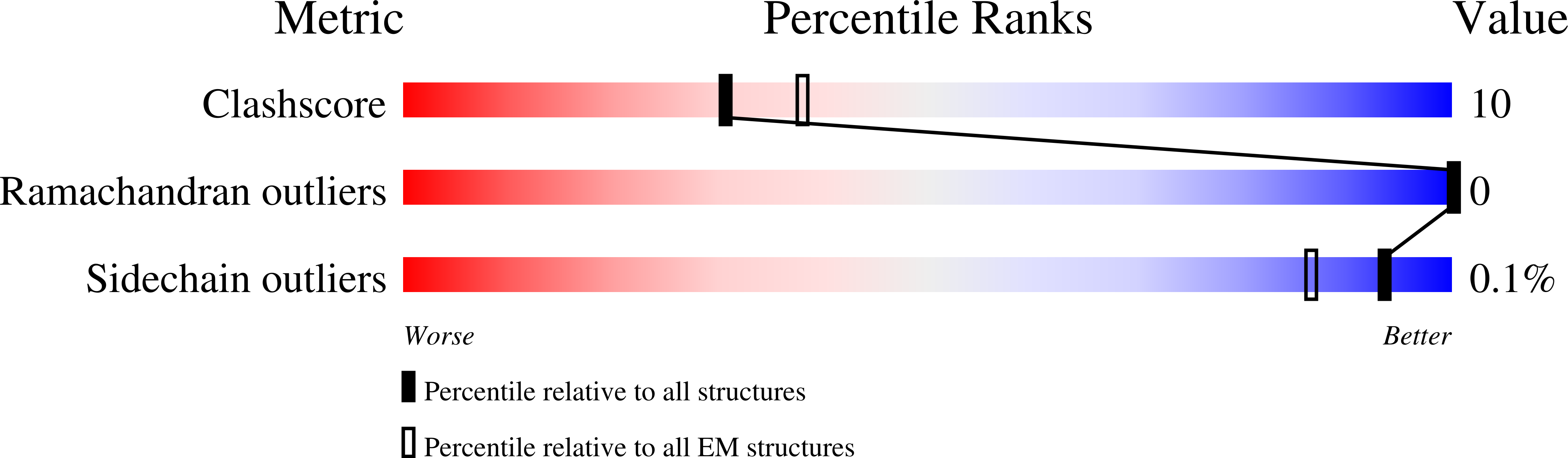Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Khare, B., Klose, T., Fang, Q., Rossmann, M.G., Kuhn, R.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34417300
- DOI: https://doi.org/10.1073/pnas.2107408118
- Primary Citation of Related Structures:
7LCG, 7LCH - PubMed Abstract:
Usutu virus (USUV) is an emerging arbovirus in Europe that has been increasingly identified in asymptomatic humans and donated blood samples and is a cause of increased incidents of neuroinvasive human disease. Treatment or prevention options for USUV disease are currently nonexistent, the result of a lack of understanding of the fundamental elements of USUV pathogenesis. Here, we report two structures of the mature USUV virus, determined at a resolution of 2.4 Å, using single-particle cryogenic electron microscopy. Mature USUV is an icosahedral shell of 180 copies of envelope (E) and membrane (M) proteins arranged in the classic herringbone pattern. However, unlike previous reports of flavivirus structures, we observe virus subpopulations and differences in the fusion loop disulfide bond. Presence of a second, unique E glycosylation site could elucidate host interactions, contributing to the broad USUV tissue tropism. The structures provide a basis for exploring USUV interactions with glycosaminoglycans and lectins, the role of the RGD motif as a receptor, and the inability of West Nile virus therapeutic antibody E16 to neutralize the mature USUV strain SAAR-1776. Finally, we identify three lipid binding sites and predict key residues that likely participate in virus stability and flexibility during membrane fusion. Our findings provide a framework for the development of USUV therapeutics and expand the current knowledge base of flavivirus biology.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.