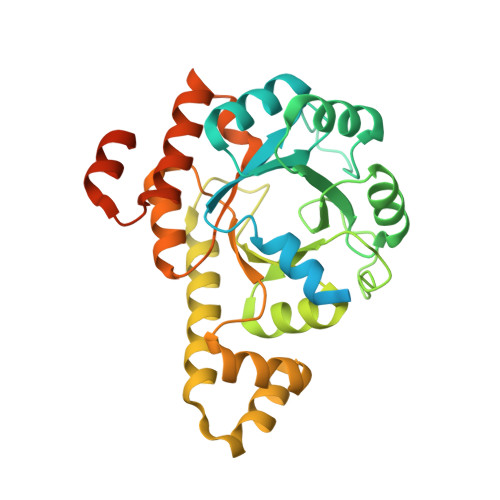
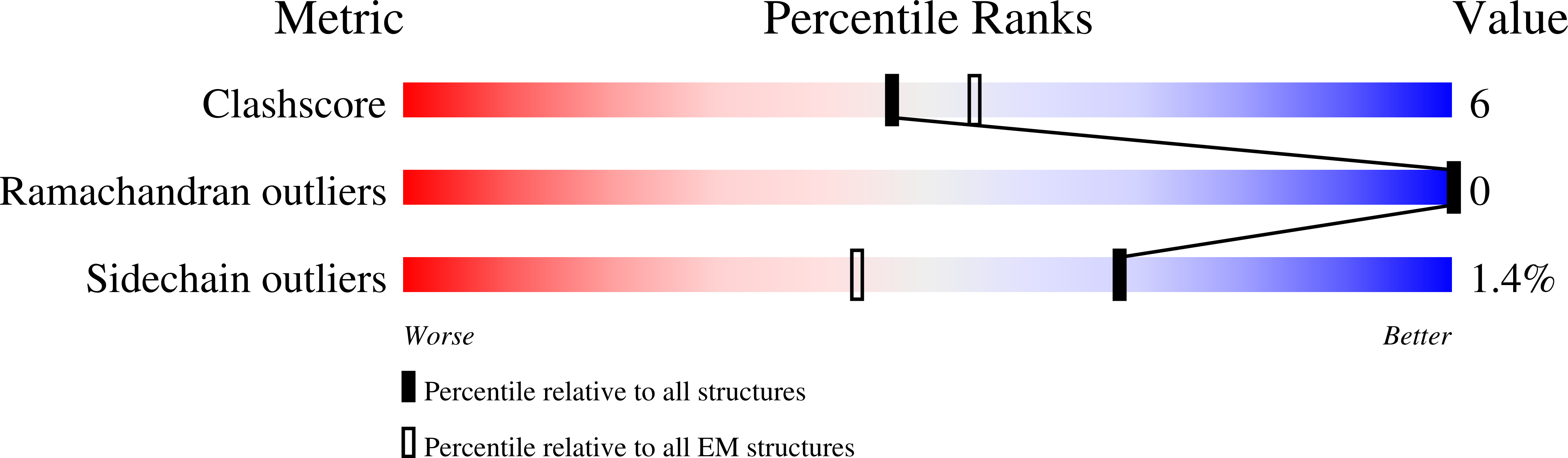Tunable Heteroassembly of a Plant Pseudoenzyme-Enzyme Complex.
Novikova, I.V., Zhou, M., Du, C., Parra, M., Kim, D.N., VanAernum, Z.L., Shaw, J.B., Hellmann, H., Wysocki, V.H., Evans, J.E.(2021) ACS Chem Biol 16: 2315-2325
- PubMed: 34520180
- DOI: https://doi.org/10.1021/acschembio.1c00475
- Primary Citation of Related Structures:
7LB5, 7LB6 - PubMed Abstract:
Pseudoenzymes have emerged as key regulatory elements in all kingdoms of life despite being catalytically nonactive. Yet many factors defining why one protein is active while its homologue is inactive remain uncertain. For pseudoenzyme-enzyme pairs, the similarity of both subunits can often hinder conventional characterization approaches. In plants, a pseudoenzyme, PDX1.2, positively regulates vitamin B 6 production by association with its active catalytic homologues such as PDX1.3 through an unknown assembly mechanism. Here we used an integrative experimental approach to learn that such pseudoenzyme-enzyme pair associations result in heterocomplexes of variable stoichiometry, which are unexpectedly tunable. We also present the atomic structure of the PDX1.2 pseudoenzyme as well as the population averaged PDX1.2-PDX1.3 pseudoenzyme-enzyme pair. Finally, we dissected hetero-dodecamers of each stoichiometry to understand the arrangement of monomers in the heterocomplexes and identified symmetry-imposed preferences in PDX1.2-PDX1.3 interactions. Our results provide a new model of pseudoenzyme-enzyme interactions and their native heterogeneity.
Organizational Affiliation:
Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99354, United States.