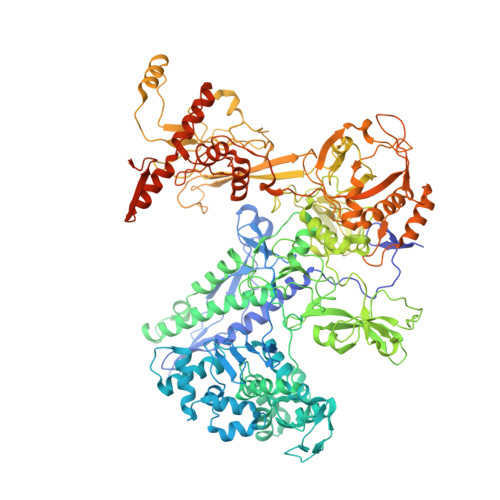
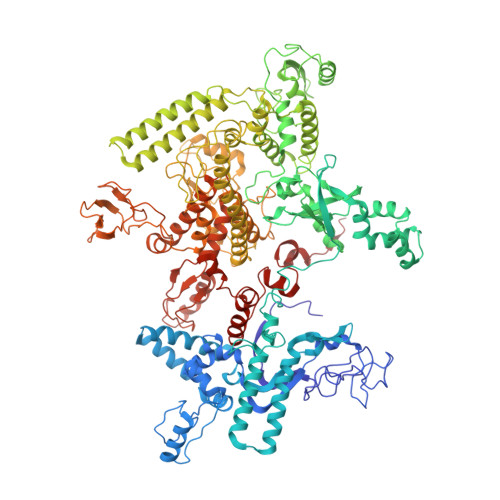
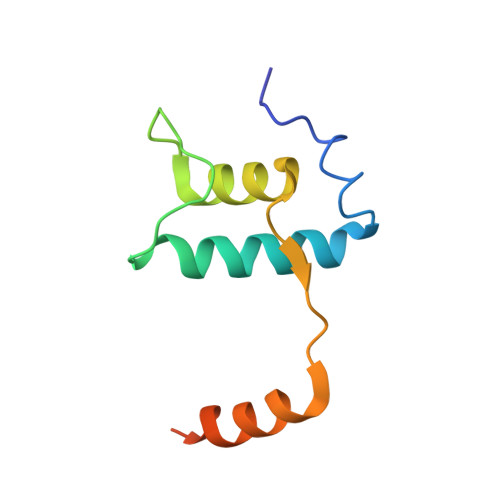
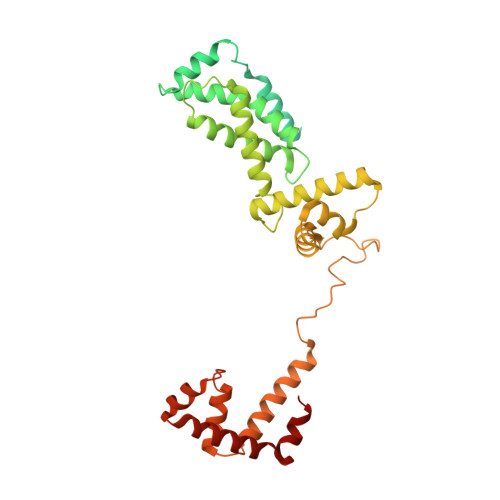
Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile.
Cao, X., Boyaci, H., Chen, J., Bao, Y., Landick, R., Campbell, E.A.(2022) Nature 604: 541-545
- PubMed: 35388215
- DOI: https://doi.org/10.1038/s41586-022-04545-z
- Primary Citation of Related Structures:
7L7B - PubMed Abstract:
Fidaxomicin (Fdx) is widely used to treat Clostridioides difficile (Cdiff) infections, but the molecular basis of its narrow-spectrum activity in the human gut microbiome remains unknown. Cdiff infections are a leading cause of nosocomial deaths 1 . Fidaxomicin, which inhibits RNA polymerase, targets Cdiff with minimal effects on gut commensals, reducing recurrence of Cdiff infection 2,3 . Here we present the cryo-electron microscopy structure of Cdiff RNA polymerase in complex with fidaxomicin and identify a crucial fidaxomicin-binding determinant of Cdiff RNA polymerase that is absent in most gut microbiota such as Proteobacteria and Bacteroidetes. By combining structural, biochemical, genetic and bioinformatic analyses, we establish that a single residue in Cdiff RNA polymerase is a sensitizing element for fidaxomicin narrow-spectrum activity. Our results provide a blueprint for targeted drug design against an important human pathogen.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, United States.