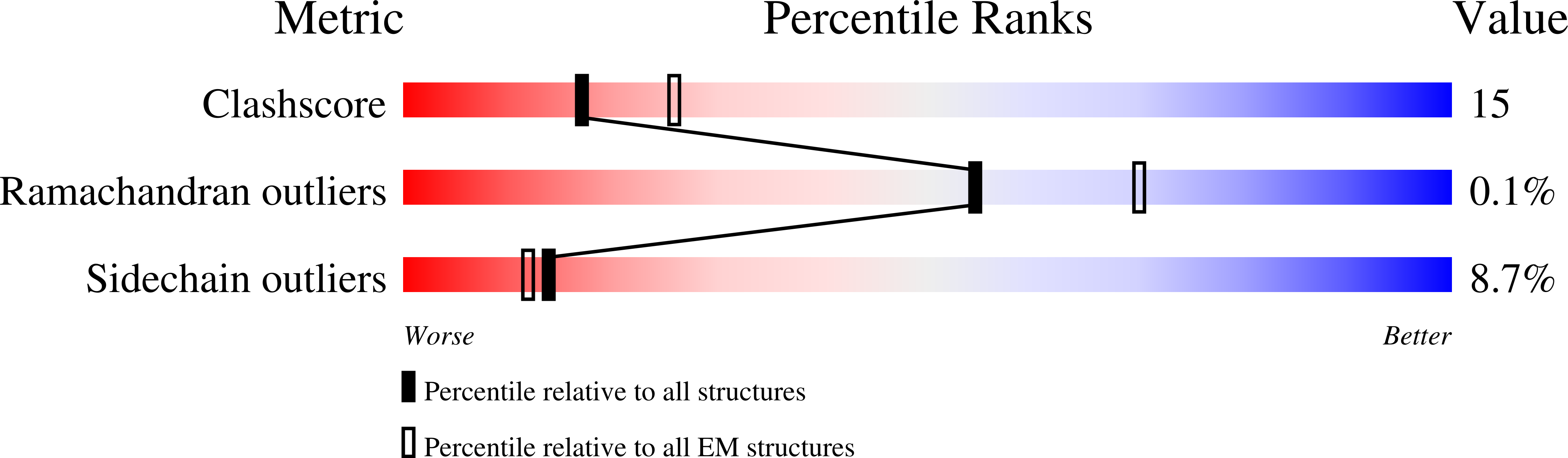A Norovirus Uses Bile Salts To Escape Antibody Recognition While Enhancing Receptor Binding.
Williams, A.N., Sherman, M.B., Smith, H.Q., Taube, S., Pettitt, B.M., Wobus, C.E., Smith, T.J.(2021) J Virol 95: e0017621-e0017621
- PubMed: 33827952
- DOI: https://doi.org/10.1128/JVI.00176-21
- Primary Citation of Related Structures:
7L5J - PubMed Abstract:
Noroviruses, members of the Caliciviridae family, are the major cause of epidemic gastroenteritis in humans, causing ∼20 million cases annually. These plus-strand RNA viruses have T=3 icosahedral protein capsids with 90 pronounced protruding (P) domain dimers to which antibodies and cellular receptors bind. In the case of mouse norovirus (MNV), bile salts have been shown to enhance receptor (CD300lf) binding to the P domain. We demonstrated previously that the P domains of several genotypes are markedly flexible and "float" over the shell, but the role of this flexibility was unclear. Recently, we demonstrated that bile causes a 90° rotation and collapse of the P domain onto the shell surface. Since bile binds distally to the P-shell interface, it was not at all clear how it could cause such dramatic changes. Here, we present the near-atomic resolution cryo-electron microscopy (cryo-EM) structure of the MNV protruding domain complexed with a neutralizing Fab. On the basis of previous results, we show here that bile salts cause allosteric conformational changes in the P domain that block antibody recognition of the top of the P domain. In addition, bile causes a major rearrangement of the P domain dimers that is likely responsible for the bile-induced collapse of the P domain onto the shell. In the contracted shell conformation, antibodies to the P1 and shell domains are not expected to bind. Therefore, at the site of infection in the gut, the host's own bile allows the virus to escape antibody-mediated neutralization while enhancing cell attachment. IMPORTANCE The major feature of calicivirus capsids is the 90 protruding domains (P domains) that are the site of cell receptor attachment and antibody epitopes. We demonstrated previously that these P domains are highly mobile and that bile causes these "floating" P domains in mouse norovirus (MNV) to contract onto the shell surface. Here, we present the near-atomic cryo-EM structure of the isolated MNV P domain complexed with a neutralizing Fab fragment. Our data show that bile causes two sets of changes. First, bile causes allosteric conformational changes in the epitopes at the top of the P domain that block antibody binding. Second, bile causes the P domain dimer subunits to rotate relative to each other, causing a contraction of the P domain that buries epitopes at the base of the P and shell domains. Taken together, the results show that MNV uses the host's own metabolites to enhance cell receptor binding while simultaneously blocking antibody recognition.
Organizational Affiliation:
University of Texas Medical Branch at Galveston, Department of Biochemistry and Molecular Biology, Galveston, Texas, USA.