Cyclobutanone Inhibitor of Cobalt-Functionalized Metallo-gamma-Lactonase AiiA with Cyclobutanone Ring Opening in the Active Site.
Reidl, C.T., Mascarenhas, R., Mohammad, T.S.H., Lutz Jr., M.R., Thomas, P.W., Fast, W., Liu, D., Becker, D.P.(2021) ACS Omega 6: 13567-13578
- PubMed: 34095651
- DOI: https://doi.org/10.1021/acsomega.0c06348
- Primary Citation of Related Structures:
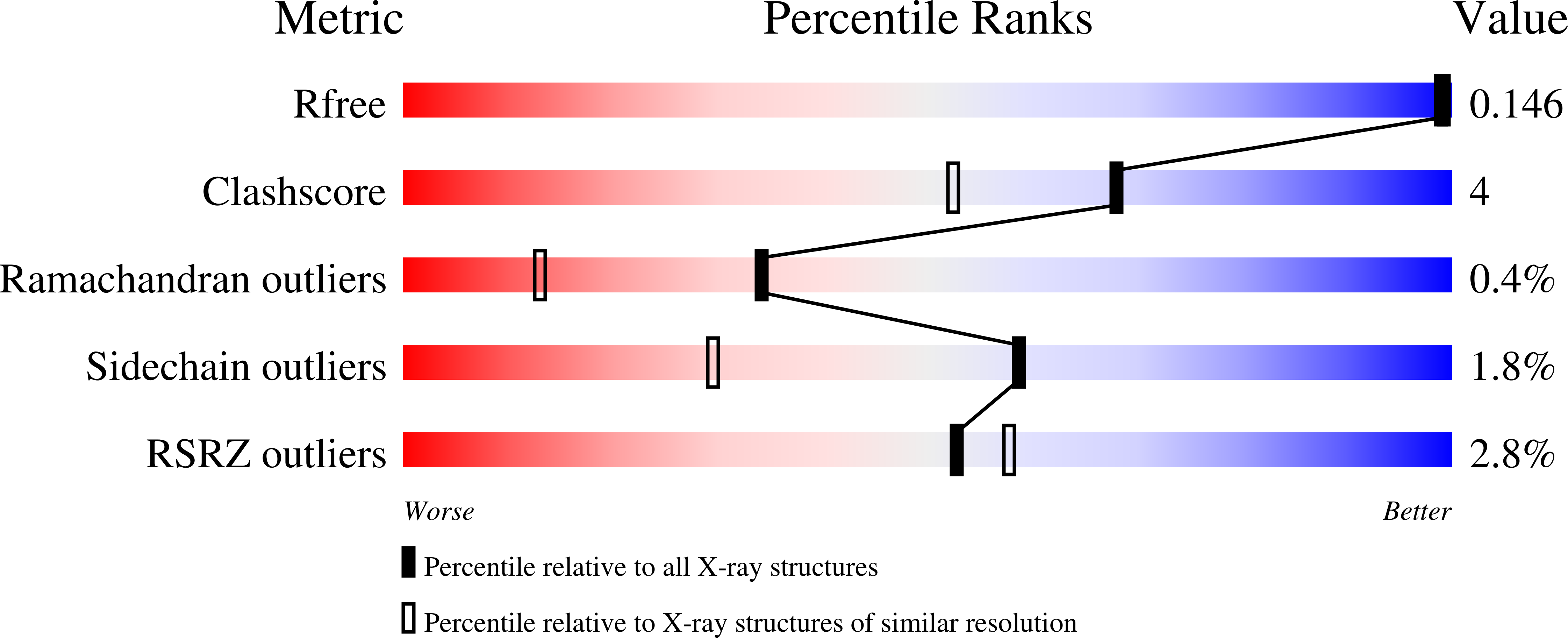
7L5F - PubMed Abstract:
An α-amido cyclobutanone possessing a C10 hydrocarbon tail was designed as a potential transition-state mimetic for the quorum-quenching metallo-γ-lactonase autoinducer inactivator A (AiiA) with the support of in-house modeling techniques and found to be a competitive inhibitor of dicobalt(II) AiiA with an inhibition constant of K i = 0.007 ± 0.002 mM. The catalytic mechanism of AiiA was further explored using our product-based transition-state modeling (PBTSM) computational approach, providing substrate-intermediate models arising during enzyme turnover and further insight into substrate-enzyme interactions governing native substrate catalysis. These interactions were targeted in the docking of cyclobutanone hydrates into the active site of AiiA. The X-ray crystal structure of dicobalt(II) AiiA cocrystallized with this cyclobutanone inhibitor unexpectedly revealed an N -(2-oxocyclobutyl)decanamide ring-opened acyclic product bound to the enzyme active site (PDB 7L5F). The C10 alkyl chain and its interaction with the hydrophobic phenylalanine clamp region of AiiA adjacent to the active site enabled atomic placement of the ligand atoms, including the C10 alkyl chain. A mechanistic hypothesis for the ring opening is proposed involving a radical-mediated process.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Loyola University Chicago, 1032 West Sheridan Road, Chicago, Illinois 60660, United States.