Identification of Potent Reverse Indazole Inhibitors for HPK1.
Yu, E.C., Methot, J.L., Fradera, X., Lesburg, C.A., Lacey, B.M., Siliphaivanh, P., Liu, P., Smith, D.M., Xu, Z., Piesvaux, J.A., Kawamura, S., Xu, H., Miller, J.R., Bittinger, M., Pasternak, A.(2021) ACS Med Chem Lett 12: 459-466
- PubMed: 33738073
- DOI: https://doi.org/10.1021/acsmedchemlett.0c00672
- Primary Citation of Related Structures:
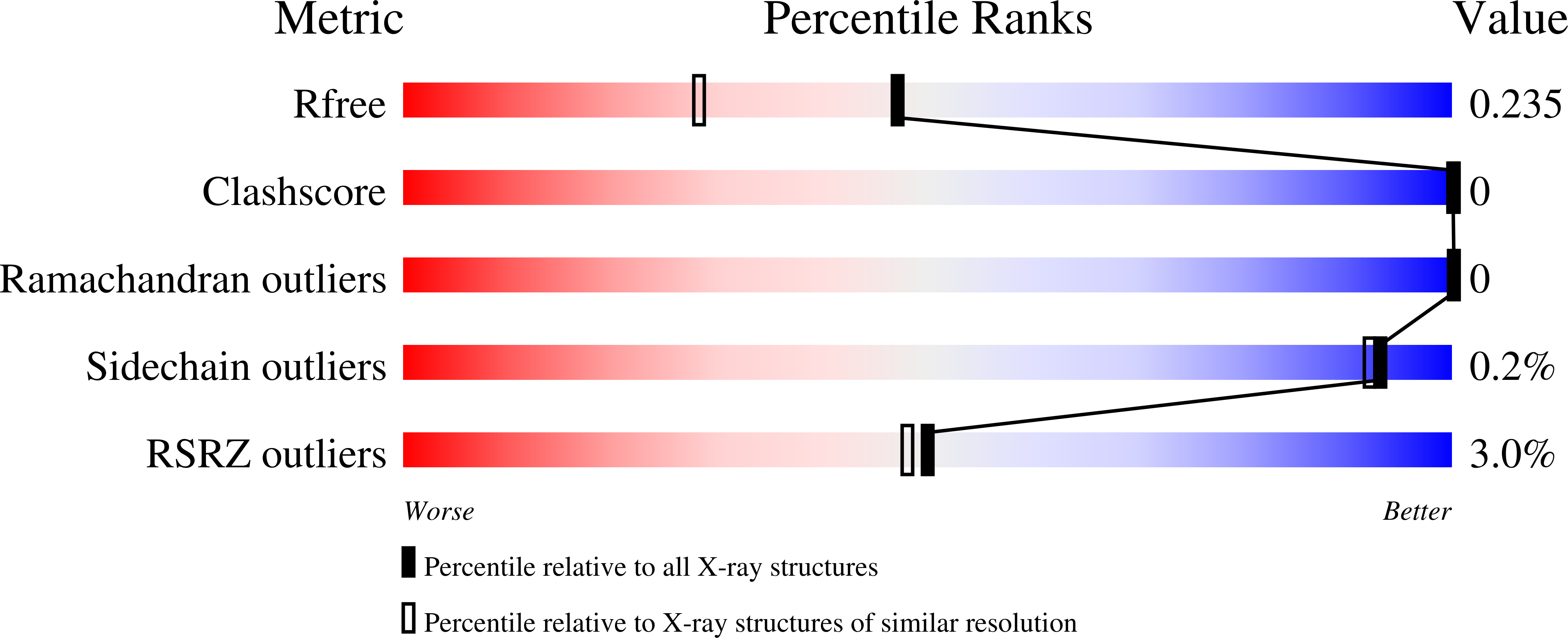
7L24, 7L25, 7L26 - PubMed Abstract:
Hematopoietic progenitor kinase (HPK1), a negative regulator of TCR-mediated T-cell activation, has been recognized as a novel antitumor immunotherapy target. Structural optimization of kinase inhibitor 4 through a systematic two-dimensional diversity screen of pyrazolopyridines led to the identification of potent and selective compounds. Crystallographic studies with HPK1 revealed a favorable water-mediated interaction with Asp155 and a salt bridge to Asp101 with optimized heterocyclic solvent fronts that were critical for enhanced potency and selectivity. Computational studies of model systems revealed differences in torsional profiles that allowed for these beneficial protein-ligand interactions. Further optimization of molecular properties led to identification of potent and selective reverse indazole inhibitor 36 that inhibited phosphorylation of adaptor protein SLP76 in human PBMC and exhibited low clearance with notable bioavailability in in vivo rat studies.
Organizational Affiliation:
Discovery Chemistry, Merck & Co., Inc., Boston, Massachusetts, 02115, United States.