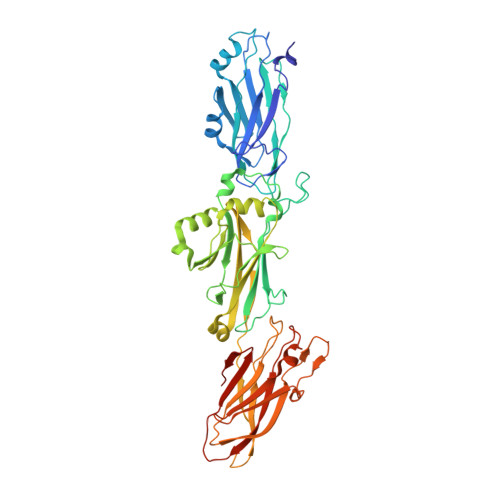
Structural and functional analysis of the C-terminal region of Streptococcus gordonii SspB.
Schormann, N., Purushotham, S., Mieher, J.L., Patel, M., Wu, H., Deivanayagam, C.(2021) Acta Crystallogr D Struct Biol 77: 1206-1215
- PubMed: 34473090
- DOI: https://doi.org/10.1107/S2059798321008135
- Primary Citation of Related Structures:
7L0O - PubMed Abstract:
Streptococcus gordonii is a member of the viridans streptococci and is an early colonizer of the tooth surface. Adherence to the tooth surface is enabled by proteins present on the S. gordonii cell surface, among which SspB belongs to one of the most well studied cell-wall-anchored adhesin families: the antigen I/II (AgI/II) family. The C-terminal region of SspB consists of three tandemly connected individual domains that display the DEv-IgG fold. These C-terminal domains contain a conserved Ca 2+ -binding site and isopeptide bonds, and they adhere to glycoprotein 340 (Gp340; also known as salivary agglutinin, SAG). Here, the structural and functional characterization of the C 123 SspB domain at 2.7 Å resolution is reported. Although the individual C-terminal domains of Streptococcus mutans AgI/II and S. gordonii SspB show a high degree of both sequence and structural homology, superposition of these structures highlights substantial differences in their electrostatic surface plots, and this can be attributed to the relative orientation of the individual domains (C 1 , C 2 and C 3 ) with respect to each other and could reflect their specificity in binding to extracellular matrix molecules. Studies further confirmed that affinity for Gp340 or its scavenger receptor cysteine-rich (SRCR) domains requires two of the three domains of C 123 SspB , namely C 12 or C 23 , which is different from AgI/II. Using protein-protein docking studies, models for this observed functional difference between C 123 SspB and C 123 AgI/II in their binding to SRCR 1 are presented.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, AL 35294, USA.