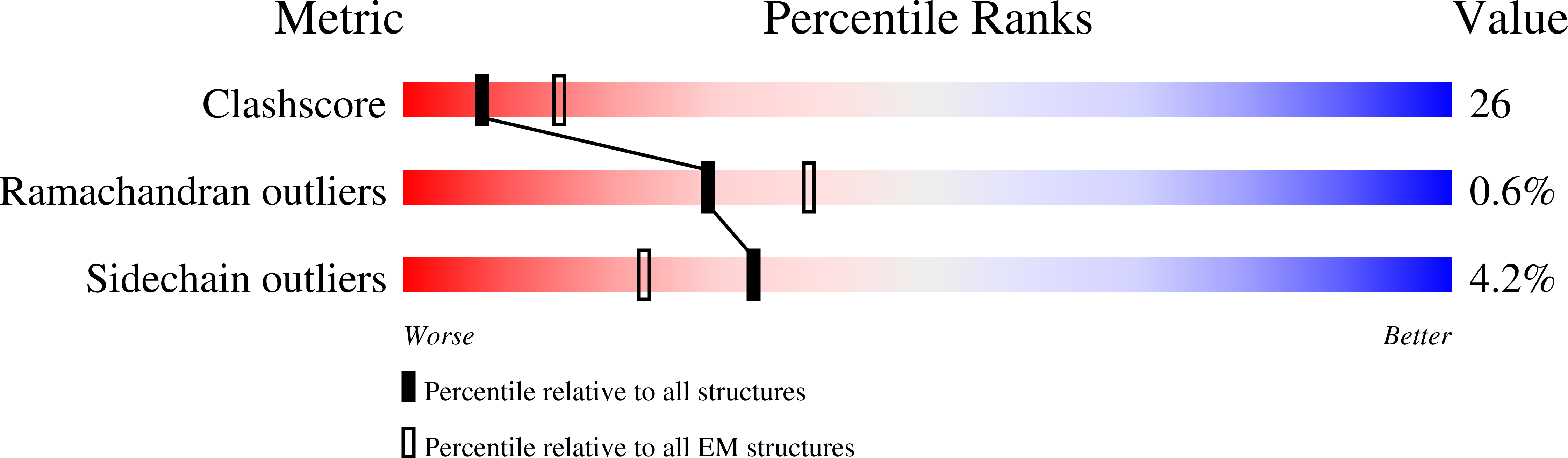Anthrax toxin translocation complex reveals insight into the lethal factor unfolding and refolding mechanism.
Machen, A.J., Fisher, M.T., Freudenthal, B.D.(2021) Sci Rep 11: 13038-13038
- PubMed: 34158520
- DOI: https://doi.org/10.1038/s41598-021-91596-3
- Primary Citation of Related Structures:
7KXR - PubMed Abstract:
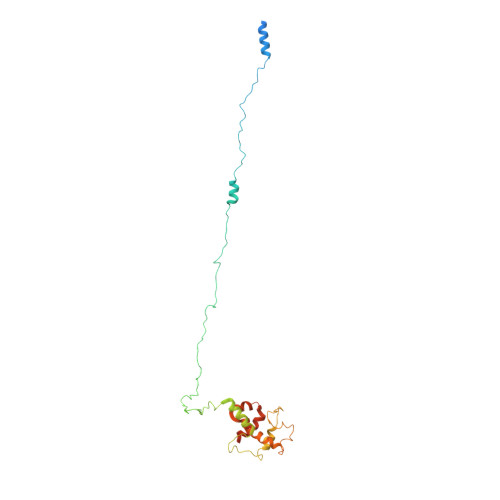
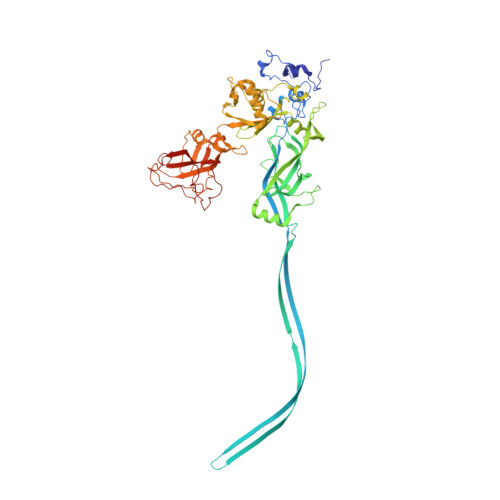
Translocation is essential to the anthrax toxin mechanism. Protective antigen (PA), the binding component of this AB toxin, forms an oligomeric pore that translocates lethal factor (LF) or edema factor, the active components of the toxin, into the cell. Structural details of the translocation process have remained elusive despite their biological importance. To overcome the technical challenges of studying translocation intermediates, we developed a method to immobilize, transition, and stabilize anthrax toxin to mimic important physiological steps in the intoxication process. Here, we report a cryoEM snapshot of PA pore translocating the N-terminal domain of LF (LF N ). The resulting 3.3 Å structure of the complex shows density of partially unfolded LF N near the canonical PA pore binding site. Interestingly, we also observe density consistent with an α helix emerging from the 100 Å β barrel channel suggesting LF secondary structural elements begin to refold in the pore channel. We conclude the anthrax toxin β barrel aids in efficient folding of its enzymatic payload prior to channel exit. Our hypothesized refolding mechanism has broader implications for pore length of other protein translocating toxins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS, 66160, USA.