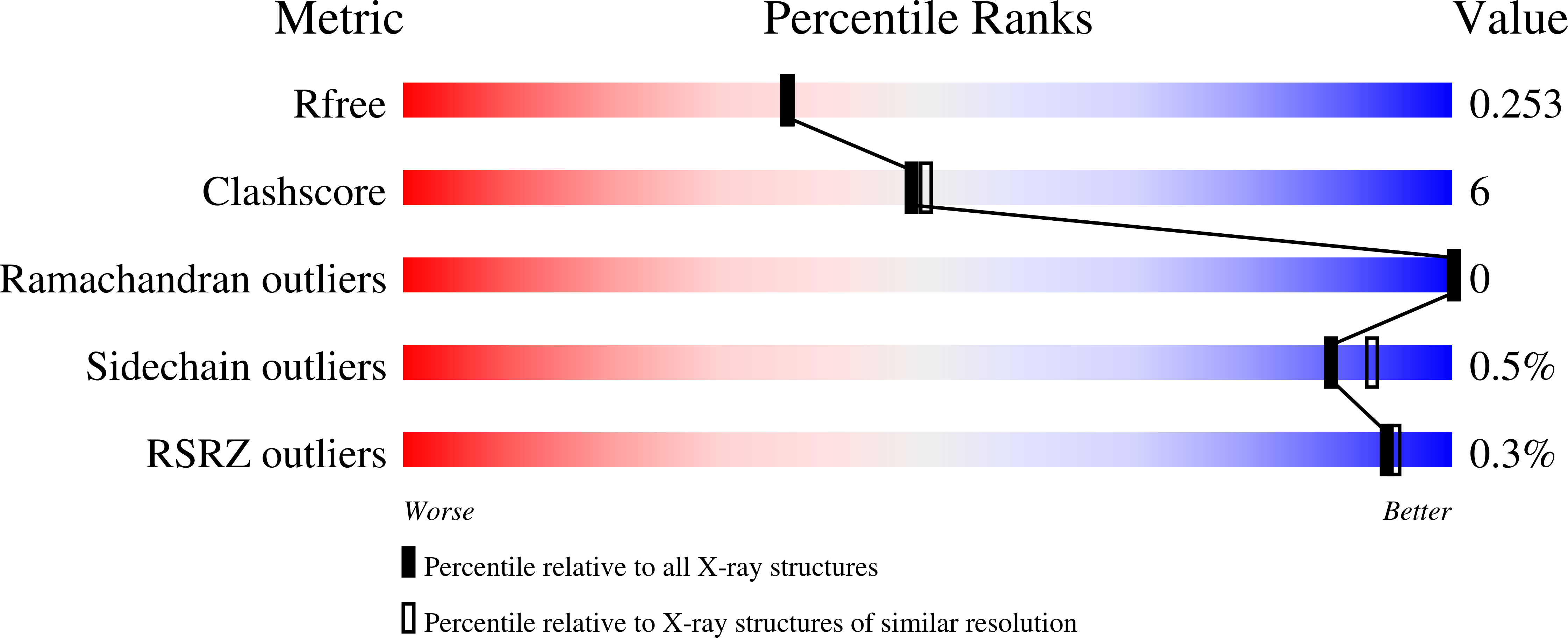
PGE (Subject of Investigation/LOI) Query on PGE Download Ideal Coordinates CCD File AB [auth E]
AB [auth E],
TRIETHYLENE GLYCOL 6 H14 O4 EDO (Subject of Investigation/LOI) Query on EDO Download Ideal Coordinates CCD File AA [auth B]
AA [auth B],
1,2-ETHANEDIOL 2 H6 O2 ACT (Subject of Investigation/LOI) Query on ACT Download Ideal Coordinates CCD File BB [auth E]
BB [auth E],
ACETATE ION 2 H3 O2