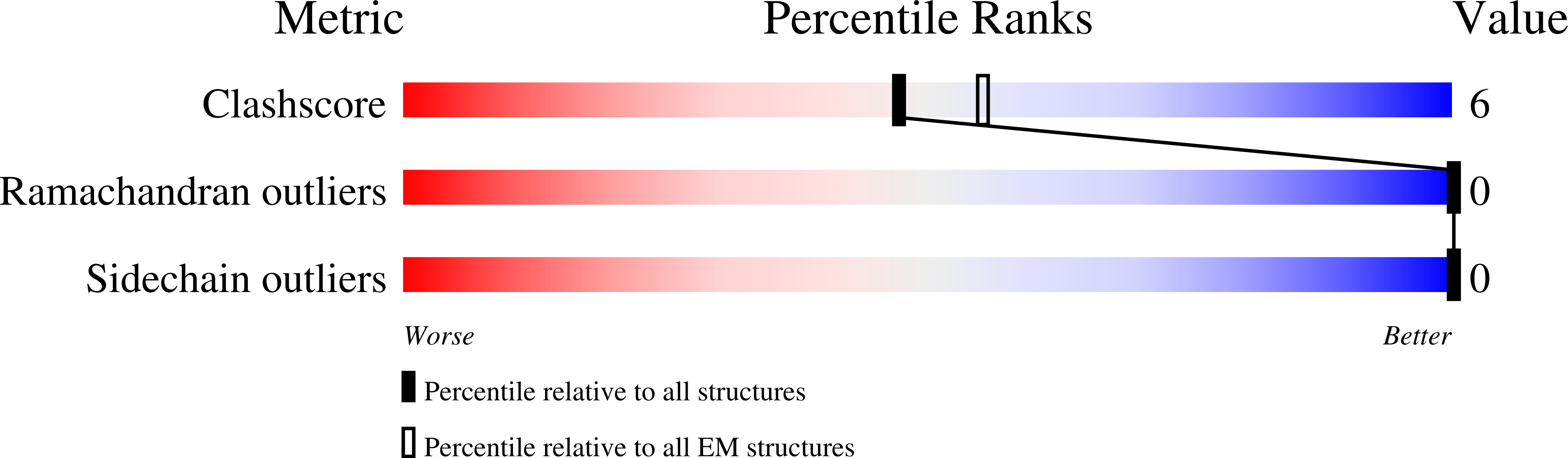Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome.
Matyszewski, M., Zheng, W., Lueck, J., Mazanek, Z., Mohideen, N., Lau, A.Y., Egelman, E.H., Sohn, J.(2021) Nat Commun 12: 2735-2735
- PubMed: 33980849
- DOI: https://doi.org/10.1038/s41467-021-23045-8
- Primary Citation of Related Structures:
7K3R - PubMed Abstract:
Inflammasomes are filamentous signaling platforms integral to innate immunity. Currently, little is known about how these structurally similar filaments recognize and distinguish one another. A cryo-EM structure of the AIM2 PYD filament reveals that the architecture of the upstream filament is essentially identical to that of the adaptor ASC PYD filament. In silico simulations using Rosetta and molecular dynamics followed by biochemical and cellular experiments consistently demonstrate that individual filaments assemble bidirectionally. By contrast, the recognition between AIM2 and ASC requires at least one to be oligomeric and occurs in a head-to-tail manner. Using in silico mutagenesis as a guide, we also identify specific axial and lateral interfaces that dictate the recognition and distinction between AIM2 and ASC filaments. Together, the results here provide a robust framework for delineating the signaling specificity and order of inflammasomes.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD, USA.