Salmonella enterica BcfH Is a Trimeric Thioredoxin-Like Bifunctional Enzyme with Both Thiol Oxidase and Disulfide Isomerase Activities.
Subedi, P., Paxman, J.J., Wang, G., Hor, L., Hong, Y., Verderosa, A.D., Whitten, A.E., Panjikar, S., Santos-Martin, C.F., Martin, J.L., Totsika, M., Heras, B.(2021) Antioxid Redox Signal 35: 21-39
- PubMed: 33607928
- DOI: https://doi.org/10.1089/ars.2020.8218
- Primary Citation of Related Structures:
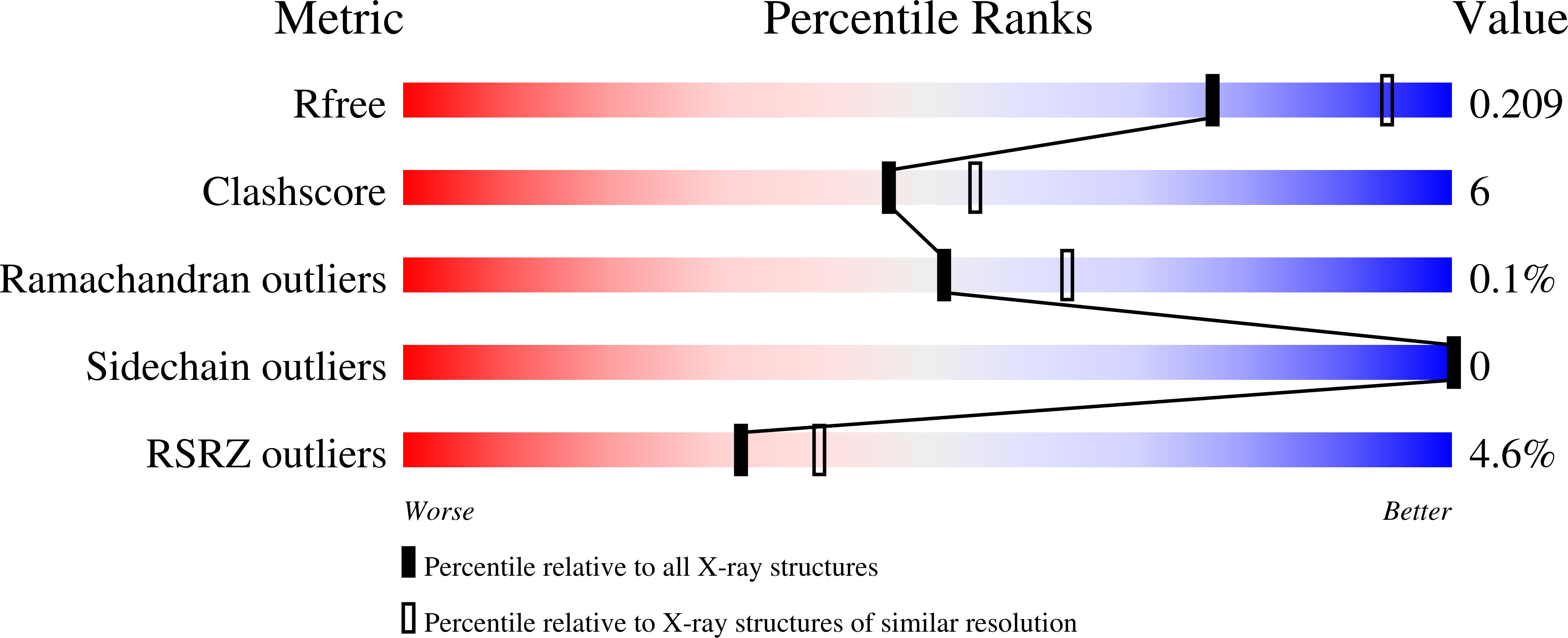
7JVE - PubMed Abstract:
Aims: Thioredoxin (TRX)-fold proteins are ubiquitous in nature. This redox scaffold has evolved to enable a variety of functions, including redox regulation, protein folding, and oxidative stress defense. In bacteria, the TRX-like disulfide bond (Dsb) family mediates the oxidative folding of multiple proteins required for fitness and pathogenic potential. Conventionally, Dsb proteins have specific redox functions with monomeric and dimeric Dsbs exclusively catalyzing thiol oxidation and disulfide isomerization, respectively. This contrasts with the eukaryotic disulfide forming machinery where the modular TRX protein disulfide isomerase (PDI) mediates thiol oxidation and disulfide reshuffling. In this study, we identified and structurally and biochemically characterized a novel Dsb-like protein from Salmonella enterica termed bovine colonization factor protein H (BcfH) and defined its role in virulence. Results: In the conserved bovine colonization factor ( bcf ) fimbrial operon, the Dsb-like enzyme BcfH forms a trimeric structure, exceptionally uncommon among the large and evolutionary conserved TRX superfamily. This protein also displays very unusual catalytic redox centers, including an unwound α-helix holding the redox active site and a trans- proline instead of the conserved cis -proline active site loop. Remarkably, BcfH displays both thiol oxidase and disulfide isomerase activities contributing to Salmonella fimbrial biogenesis. Innovation and Conclusion: Typically, oligomerization of bacterial Dsb proteins modulates their redox function, with monomeric and dimeric Dsbs mediating thiol oxidation and disulfide isomerization, respectively. This study demonstrates a further structural and functional malleability in the TRX-fold protein family. BcfH trimeric architecture and unconventional catalytic sites permit multiple redox functions emulating in bacteria the eukaryotic PDI dual oxidoreductase activity. Antioxid. Redox Signal . 35, 21-39.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Bundoora, Australia.