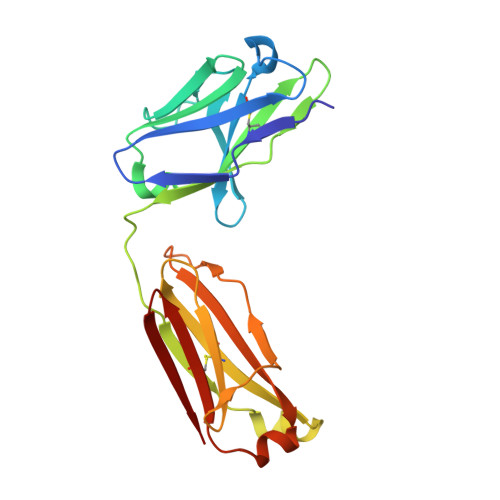
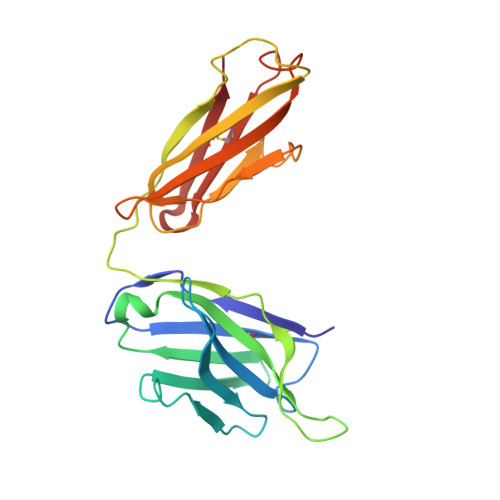
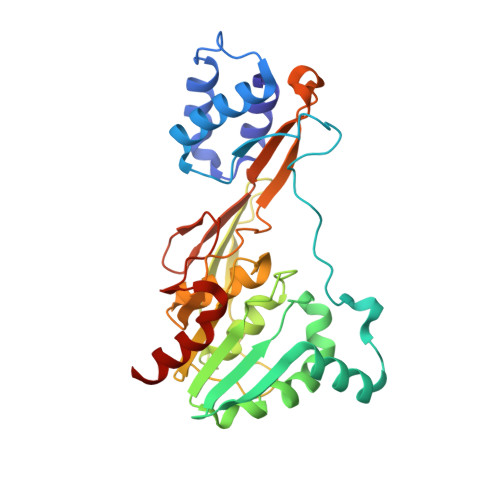
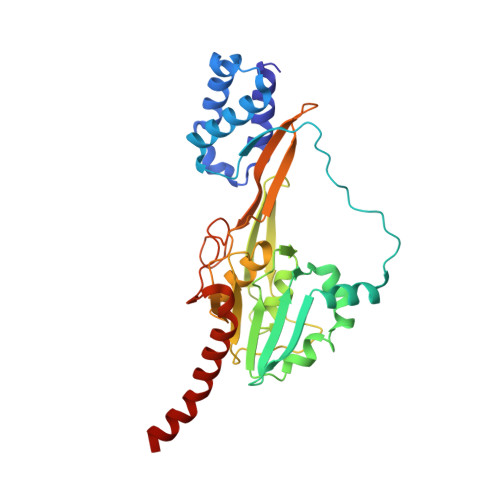
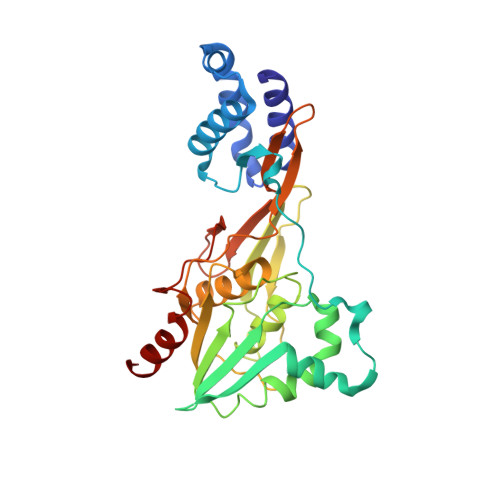
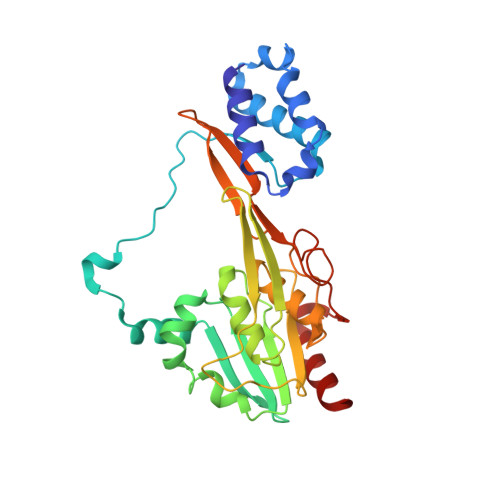
Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody.
Sears, A.E., Albiez, S., Gulati, S., Wang, B., Kiser, P., Kovacik, L., Engel, A., Stahlberg, H., Palczewski, K.(2020) FASEB J 34: 13918-13934
- PubMed: 32860273
- DOI: https://doi.org/10.1096/fj.202000796RR
- Primary Citation of Related Structures:
7JTI - PubMed Abstract:
Interphotoreceptor retinoid-binding protein (IRBP) is a highly expressed protein secreted by rod and cone photoreceptors that has major roles in photoreceptor homeostasis as well as retinoid and polyunsaturated fatty acid transport between the neural retina and retinal pigment epithelium. Despite two crystal structures reported on fragments of IRBP and decades of research, the overall structure of IRBP and function within the visual cycle remain unsolved. Here, we studied the structure of native bovine IRBP in complex with a monoclonal antibody (mAb5) by cryo-electron microscopy, revealing the tertiary and quaternary structure at sufficient resolution to clearly identify the complex components. Complementary mass spectrometry experiments revealed the structure and locations of N-linked carbohydrate post-translational modifications. This work provides insight into the structure of IRBP, displaying an elongated, flexible three-dimensional architecture not seen among other retinoid-binding proteins. This work is the first step in elucidation of the function of this enigmatic protein.
Organizational Affiliation:
Department of Pharmacology, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.