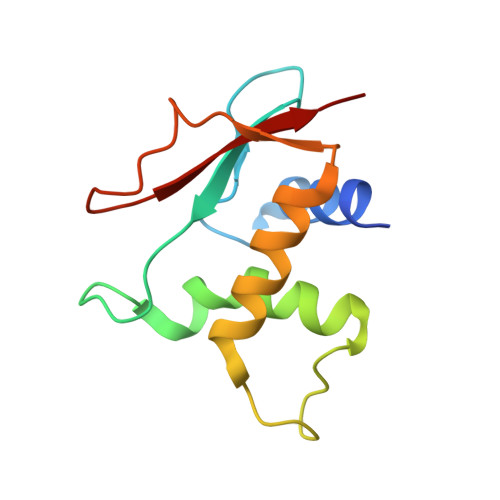
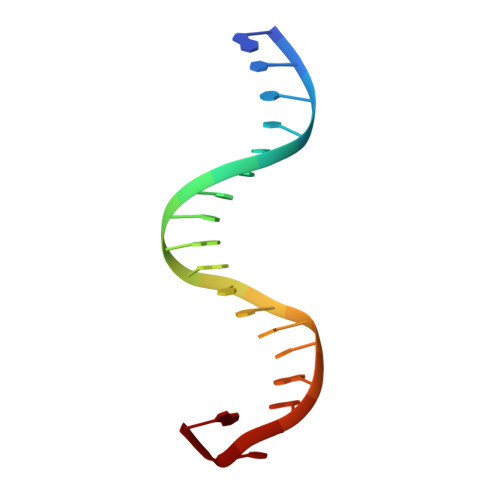
Structural determinants of the IRF4/DNA homodimeric complex.
Sundararaj, S., Seneviratne, S., Williams, S.J., Enders, A., Casarotto, M.G.(2021) Nucleic Acids Res 49: 2255-2265
- PubMed: 33533913
- DOI: https://doi.org/10.1093/nar/gkaa1287
- Primary Citation of Related Structures:
7JM4 - PubMed Abstract:
Interferon regulatory factor 4 (IRF4) is a key transcription factor (TF) in the regulation of immune cells, including B and T cells. It acts by binding DNA as both a homodimer and, in conjunction with other TFs, as a heterodimer. The choice of homo and heterodimeric/ DNA interactions is a critical aspect in the control of the transcriptional program and cell fate outcome. To characterize the nature of this interaction in the homodimeric complex, we have determined the crystal structure of the IRF4/ISRE homodimeric complex. We show that the complex formation is aided by a substantial DNA deformation with co-operative binding achieved exclusively through protein-DNA contact. This markedly contrasts with the heterodimeric form where DNA bound IRF4 is shown to physically interact with PU.1 TF to engage EICE1. We also show that the hotspot residues (Arg98, Cys99 and Asn102) contact both consensus and non-consensus sequences with the L1 loop exhibiting marked flexibility. Additionally, we identified that IRF4L116R, a mutant associated with chronic lymphocytic leukemia, binds more robustly to DNA thereby providing a rationale for the observed gain of function. Together, we demonstrate key structural differences between IRF4 homo and heterodimeric complexes, thereby providing molecular insights into IRF4-mediated transcriptional regulation.
Organizational Affiliation:
Eccles Institute of Neuroscience, John Curtin School of Medical Research, Australian National University, Canberra 2600, Australia.