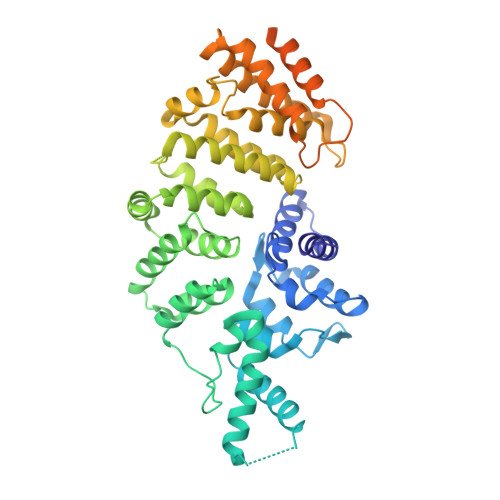
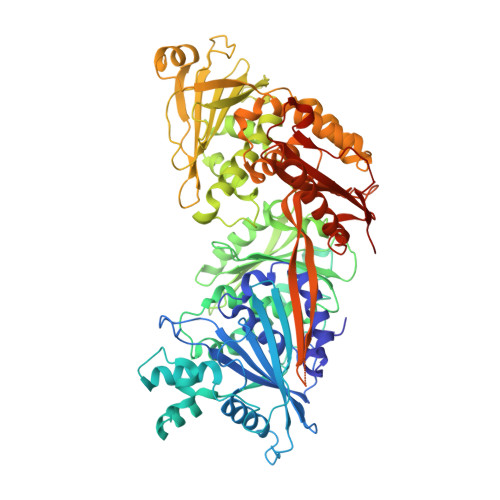
Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Wang, H., Xiao, Y., Chen, X., Zhang, M., Sun, G., Wang, F., Wang, L., Zhang, H., Zhang, X., Yang, X., Li, W., Wei, Y., Yao, D., Zhang, B., Li, J., Cui, W., Wang, F., Chen, C., Shen, W., Su, D., Bai, F., Huang, J., Ye, S., Zhang, L., Ji, X., Wang, W., Wang, Z., Hochstrasser, M., Yang, H.(2022) Nat Commun 13: 1608-1608
- PubMed: 35338130
- DOI: https://doi.org/10.1038/s41467-022-29273-w
- Primary Citation of Related Structures:
7FIT, 7FIU, 7FIV, 7FIW - PubMed Abstract:
Cytoplasmic incompatibility (CI) results when Wolbachia bacteria-infected male insects mate with uninfected females, leading to embryonic lethality. "Rescue" of viability occurs if the female harbors the same Wolbachia strain. CI is caused by linked pairs of Wolbachia genes called CI factors (CifA and CifB). The co-evolution of CifA-CifB pairs may account in part for the incompatibility patterns documented in insects infected with different Wolbachia strains, but the molecular mechanisms remain elusive. Here, we use X-ray crystallography and AlphaFold to analyze the CI factors from Wolbachia strain wMel called CidA wMel and CidB wMel . Substituting CidA wMel interface residues with those from CidA wPip (from strain wPip) enables the mutant protein to bind CidB wPip and rescue CidB wPip -induced yeast growth defects, supporting the importance of CifA-CifB interaction in CI rescue. Sequence divergence in CidA wPip and CidB wPip proteins affects their pairwise interactions, which may help explain the complex incompatibility patterns of mosquitoes infected with different wPip strains.
Organizational Affiliation:
School of Life Sciences, Tianjin University, Tianjin, China.