Structural basis for peptide recognition by archaeal oligopeptide permease A.
Yokoyama, H., Kamei, N., Konishi, K., Hara, K., Ishikawa, Y., Matsui, I., Forterre, P., Hashimoto, H.(2022) Proteins 90: 1434-1442
- PubMed: 35170084
- DOI: https://doi.org/10.1002/prot.26324
- Primary Citation of Related Structures:
7FI3 - PubMed Abstract:
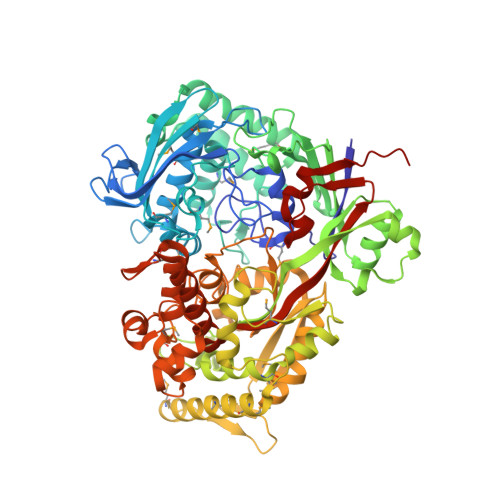
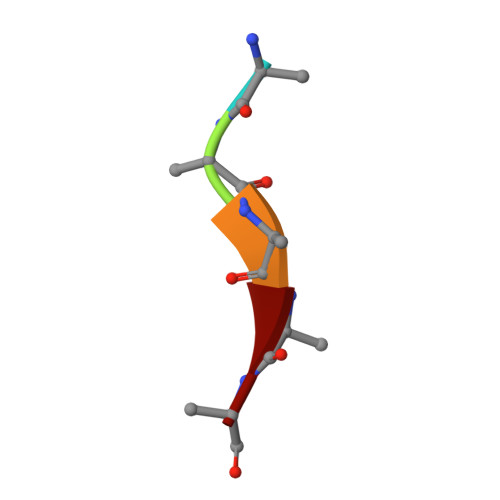
Oligopeptide permease A (OppA) plays an important role in the nutrition of cells and various signaling processes. In archaea, OppA is a major protein present in membrane vesicles of Thermococcales. Because there being no crystal structures of archaeal OppAs determined to date, we report the crystal structure of archaeal OppA from Thermococcus kodakaraensis (TkOppA) at 2.3 Å resolution by the single-wavelength anomalous dispersion method. TkOppA consists of three domains similarly to bacterial OppAs, and the inserted regions not present in bacterial OppAs are at the periphery of the core region. An endogenous pentapeptide was bound in the pocket of domains I and III of TkOppA by hydrogen bonds of main-chain atoms of the peptide and hydrophobic interactions. No hydrogen bonds of side-chain atoms of the peptide were observed; thus, TkOppA may have low peptide selectivity but some preference for residues 2 and 3. TkOppA has a relatively large pocket and can bind a nonapeptide; therefore, it is suitable for the binding of large peptides similarly to OppAs of Gram-positive bacteria.
Organizational Affiliation:
Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda, Chiba, Japan.