Structural insight into dynamic characteristic of the polymerized forms of kinetoplastid membrane protein-11
Mo, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Kinetoplastid membrane protein KMP-11 | 92 | Trypanosoma brucei gambiense DAL972 | Mutation(s): 0 Gene Names: TbgDal_IX8640, TbgDal_IX8650, TbgDal_IX8660, TbgDal_IX8670 | 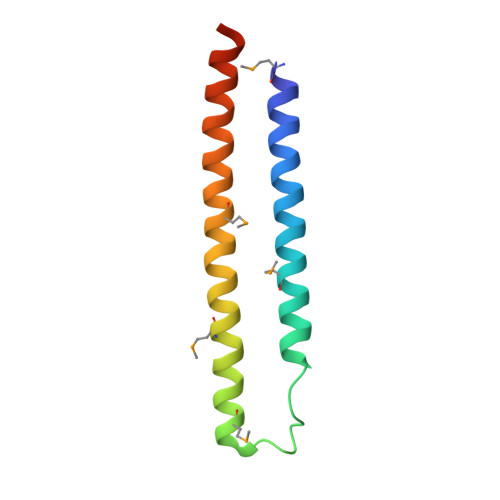 | |
UniProt | |||||
Find proteins for C9ZZD9 (Trypanosoma brucei gambiense (strain MHOM/CI/86/DAL972)) Explore C9ZZD9 Go to UniProtKB: C9ZZD9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C9ZZD9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 34.899 | α = 90 |
| b = 38.544 | β = 90 |
| c = 142.095 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32071476 |