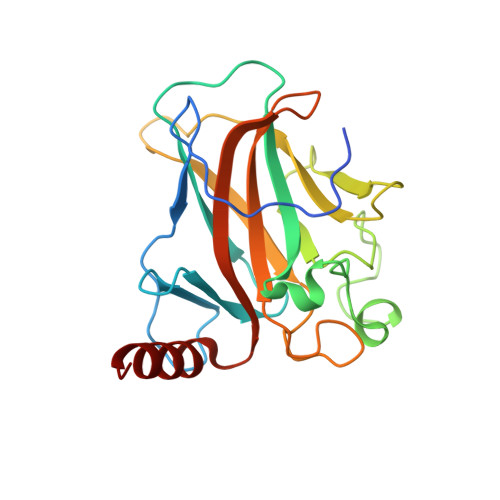
Deciphering the mechanism of p73 recognition of p53 response elements using the crystal structure of p73-DNA complexes and computational studies.
Koley, T., Roy Chowdhury, S., Kushwaha, T., Kumar, M., Inampudi, K.K., Kaur, P., Singh, T.P., Viadiu, H., Ethayathulla, A.S.(2022) Int J Biol Macromol 206: 40-50
- PubMed: 35217090
- DOI: https://doi.org/10.1016/j.ijbiomac.2022.02.108
- Primary Citation of Related Structures:
7EZJ - PubMed Abstract:
p73 belongs to p53 family transcription factor activating more than 50% of cell fate p53 target genes involved in cell cycle, apoptosis, DNA damage response alongside neuronal system development and differentiation by binding to 20-bp response elements (REs) having sequence motif (PPPC-A/T-T/A-GYYY) where P-purines and Y-pyrimidines with each 10-bp separated by minimum 0 to 13-bp spacer. The promiscuous nature of recognizing both cell fate and development genes and the underlying RE selectivity mechanism by p73 is not well understood. Here, we report the molecular details of p73 recognizing the REs using the crystal structure of p73 DNA binding domain (DBD) in complex with 12 base pair DNA sequence 5'-cAGGCATGCCTg-3' and molecular dynamics simulations with six different p53 natural promoter sequences. Each 20-base pair natural promoter forms a different major/minor groove due to the presence of nucleotides A/T, A/C, G/G, T/T and G/T at positions 3, 8, 13, 18 uniquely recognized by p73 key residues Lys138 and Arg268. The loops L1 and L3 bearing these residues influence inter-and intra-dimer interfaces interactions and hence p73 forms a unique tetramer with each natural promoter sequence. Structural features of the DNA and the spacing between half-sites influence p73 tetramerization and its transactivation function.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110029, India.