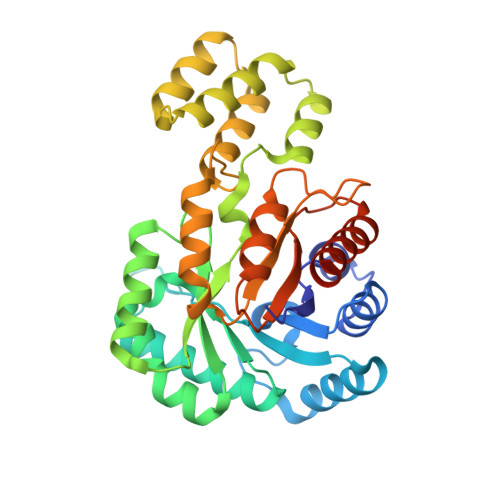
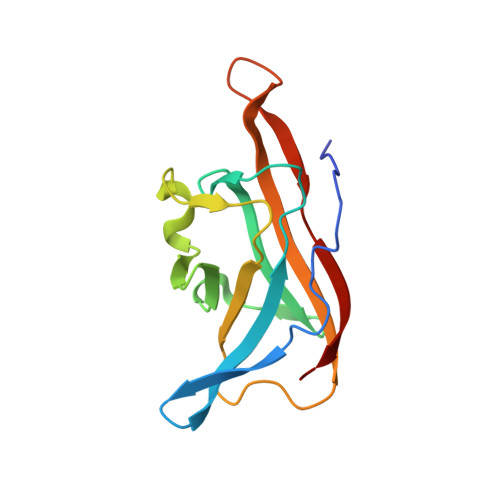
C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Mori, T., Kumano, T., He, H., Watanabe, S., Senda, M., Moriya, T., Adachi, N., Hori, S., Terashita, Y., Kawasaki, M., Hashimoto, Y., Awakawa, T., Senda, T., Abe, I., Kobayashi, M.(2021) Nat Commun 12: 6294-6294
- PubMed: 34728636
- DOI: https://doi.org/10.1038/s41467-021-26585-1
- Primary Citation of Related Structures:
7BVR, 7BVS, 7DRD, 7DRE, 7EXB, 7EXZ - PubMed Abstract:
C-Glycosides, in which a sugar moiety is linked via a carbon-carbon (C-C) bond to a non-sugar moiety (aglycone), are found in our food and medicine. The C-C bond is cleaved by intestinal microbes and the resulting aglycones exert various bioactivities. Although the enzymes responsible for the reactions have been identified, their catalytic mechanisms and the generality of the reactions in nature remain to be explored. Here, we present the identification and structural basis for the activation of xenobiotic C-glycosides by heterocomplex C-deglycosylation enzymes from intestinal and soil bacteria. They are found to be metal-dependent enzymes exhibiting broad substrate specificity toward C-glycosides. X-ray crystallographic and cryo-electron microscopic analyses, as well as structure-based mutagenesis, reveal the structural details of these enzymes and the detailed catalytic mechanisms of their remarkable C-C bond cleavage reactions. Furthermore, bioinformatic and biochemical analyses suggest that the C-deglycosylation enzymes are widely distributed in the gut, soil, and marine bacteria.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan.