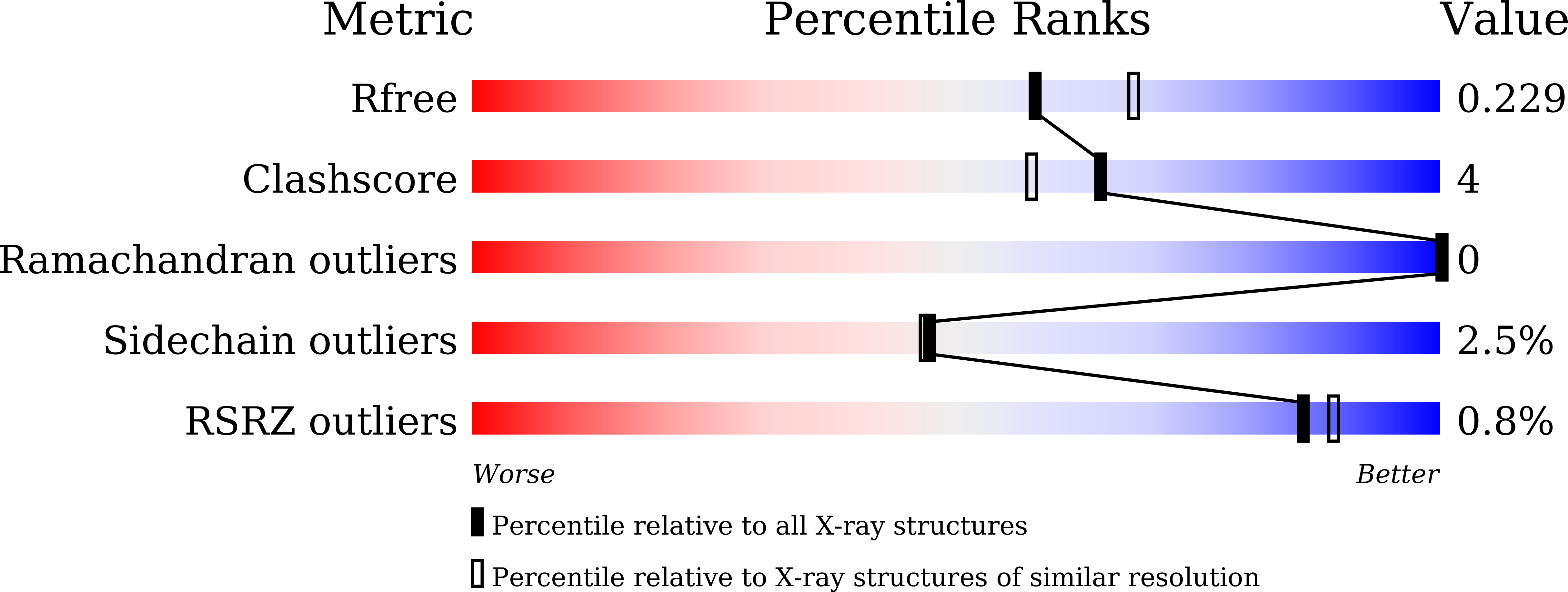
EDO Query on EDO Download Ideal Coordinates CCD File AA [auth B]
AA [auth B],
1,2-ETHANEDIOL 2 H6 O2