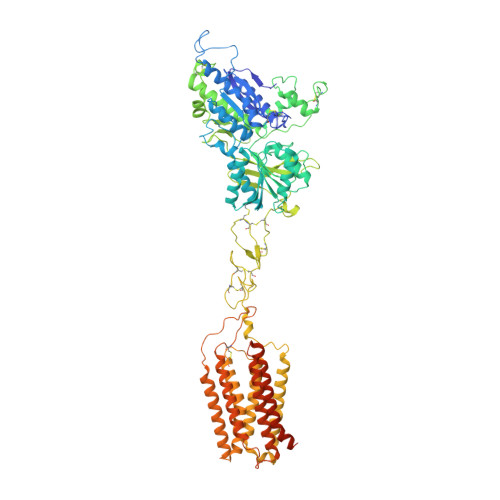
Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Du, J., Wang, D., Fan, H., Xu, C., Tai, L., Lin, S., Han, S., Tan, Q., Wang, X., Xu, T., Zhang, H., Chu, X., Yi, C., Liu, P., Wang, X., Zhou, Y., Pin, J.P., Rondard, P., Liu, H., Liu, J., Sun, F., Wu, B., Zhao, Q.(2021) Nature 594: 589-593
- PubMed: 34135509
- DOI: https://doi.org/10.1038/s41586-021-03641-w
- Primary Citation of Related Structures:
7EPA, 7EPB, 7EPC, 7EPD, 7EPE, 7EPF - PubMed Abstract:
The metabotropic glutamate receptors (mGlus) are involved in the modulation of synaptic transmission and neuronal excitability in the central nervous system 1 . These receptors probably exist as both homo- and heterodimers that have unique pharmacological and functional properties 2-4 . Here we report four cryo-electron microscopy structures of the human mGlu subtypes mGlu2 and mGlu7, including inactive mGlu2 and mGlu7 homodimers; mGlu2 homodimer bound to an agonist and a positive allosteric modulator; and inactive mGlu2-mGlu7 heterodimer. We observed a subtype-dependent dimerization mode for these mGlus, as a unique dimer interface that is mediated by helix IV (and that is important for limiting receptor activity) exists only in the inactive mGlu2 structure. The structures provide molecular details of the inter- and intra-subunit conformational changes that are required for receptor activation, which distinguish class C G-protein-coupled receptors from those in classes A and B. Furthermore, our structure and functional studies of the mGlu2-mGlu7 heterodimer suggest that the mGlu7 subunit has a dominant role in controlling dimeric association and G-protein activation in the heterodimer. These insights into mGlu homo- and heterodimers highlight the complex landscape of mGlu dimerization and activation.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.