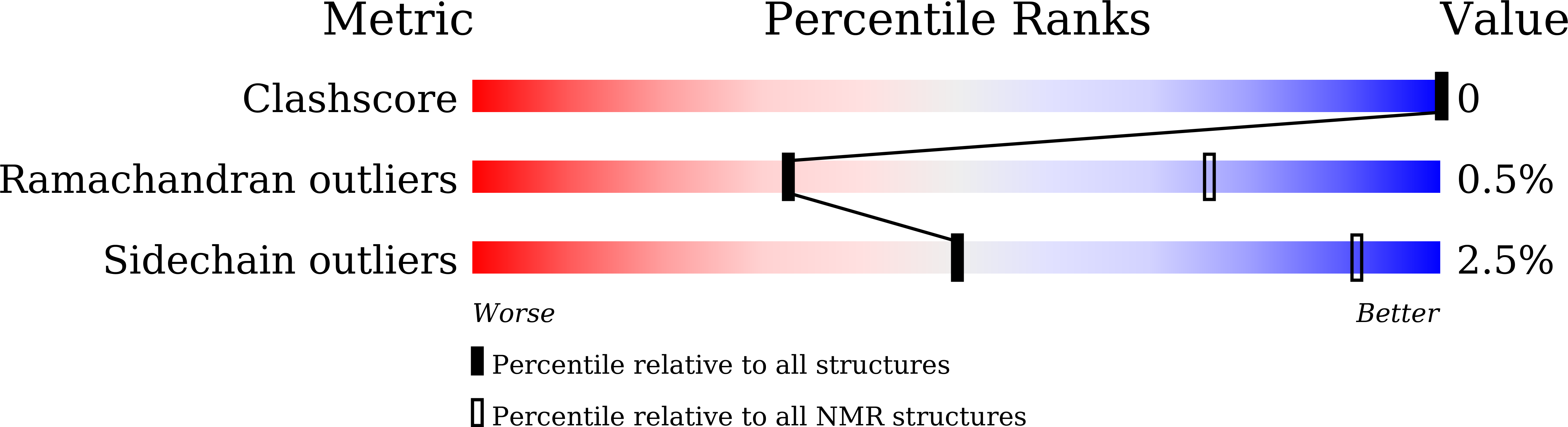
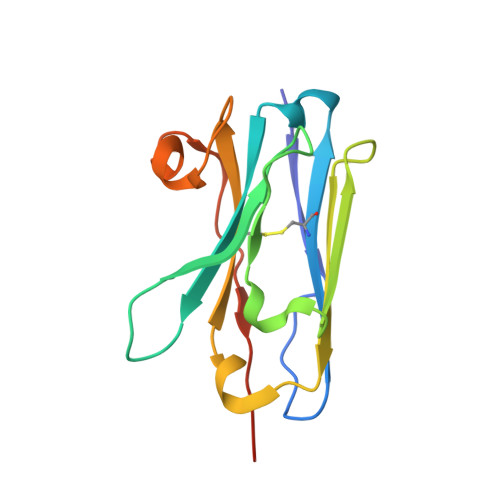
Structure of Nanobody Nb23.
Percipalle, M., Hunashal, Y., Steyaert, J., Fogolari, F., Esposito, G.(2021) Molecules 26
- PubMed: 34207949
- DOI: https://doi.org/10.3390/molecules26123567
- Primary Citation of Related Structures:
7EH3 - PubMed Abstract:
Nanobodies, or VHHs, are derived from heavy chain-only antibodies (hcAbs) found in camelids. They overcome some of the inherent limitations of monoclonal antibodies (mAbs) and derivatives thereof, due to their smaller molecular size and higher stability, and thus present an alternative to mAbs for therapeutic use. Two nanobodies, Nb23 and Nb24, have been shown to similarly inhibit the self-aggregation of very amyloidogenic variants of β2-microglobulin. Here, the structure of Nb23 was modeled with the Chemical-Shift (CS)-Rosetta server using chemical shift assignments from nuclear magnetic resonance (NMR) spectroscopy experiments, and used as prior knowledge in PONDEROSA restrained modeling based on experimentally assessed internuclear distances. Further validation was comparatively obtained with the results of molecular dynamics trajectories calculated from the resulting best energy-minimized Nb23 conformers. 2D and 3D NMR spectroscopy experiments were carried out to determine the assignment of the backbone and side chain hydrogen, nitrogen and carbon resonances to extract chemical shifts and interproton separations for restrained modeling. The solution structure of isolated Nb23 nanobody was determined. The structural analysis indicated that isolated Nb23 has a dynamic CDR3 loop distributed over different orientations with respect to Nb24, which could determine differences in target antigen affinity or complex lability.
Organizational Affiliation:
Science Division, New York University Abu Dhabi, Abu Dhabi 129188, United Arab Emirates.