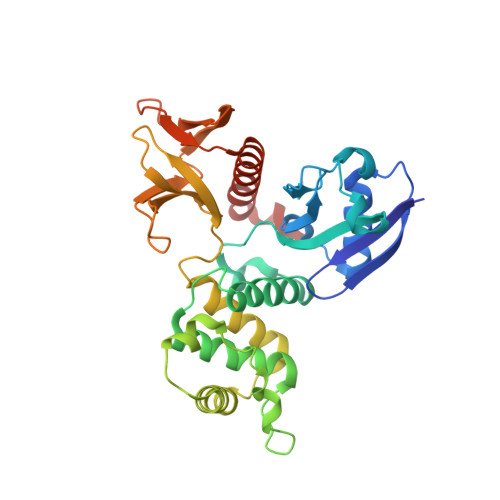
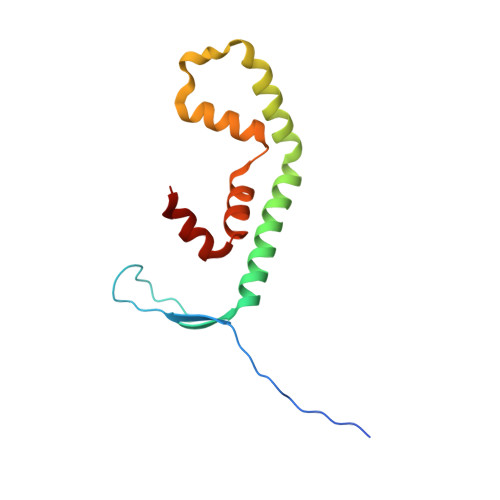
The crystal structure of the FERM and C-terminal domain complex of Drosophila Merlin.
Zhang, F., Liu, B., Gao, Y., Long, J., Zhou, H.(2021) Biochem Biophys Res Commun 553: 92-98
- PubMed: 33765559
- DOI: https://doi.org/10.1016/j.bbrc.2021.03.065
- Primary Citation of Related Structures:
7EDR - PubMed Abstract:
NF2/Merlin is an upstream regulator of hippo pathway, and it has two states: an auto-inhibited "closed" state and an active "open" form. Previous studies showed that Drosophila Merlin adopts a more closed conformation. However, the molecular mechanism of conformational regulation remains poorly understood. Here, we first confirmed the strong interaction between FERM and the C-terminal domain (CTD) of Merlin, and then determined the crystal structure of the FERM/CTD complex, which reveals the structural basis of Merlin adopting a more closed conformation compared to its human cognate NF2. Interestingly, we found that the conserved lipid-binding site of Merlin might be masked by a linker. Confocal analyses confirmed that all putative lipid-binding site are very important for the membranal location of Merlin. More, we found that the phosphomimic Thr616Asp mutation weakens the interaction between FERM and CTD of Merlin. Collectively, the crystal structure of the FERM/CTD complex not only provides a mechanistic explanation of functionally dormant conformation of Merlin may also serve as a foundation for revealing the mechanism of conformational regulation of Merlin.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, Tianjin Key Laboratory of Protein Science, College of Life Sciences, Nankai University, 94 Weijin Road, Tianjin, 300071, China.