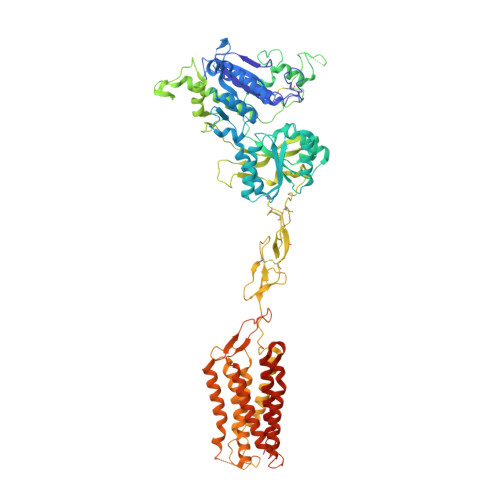
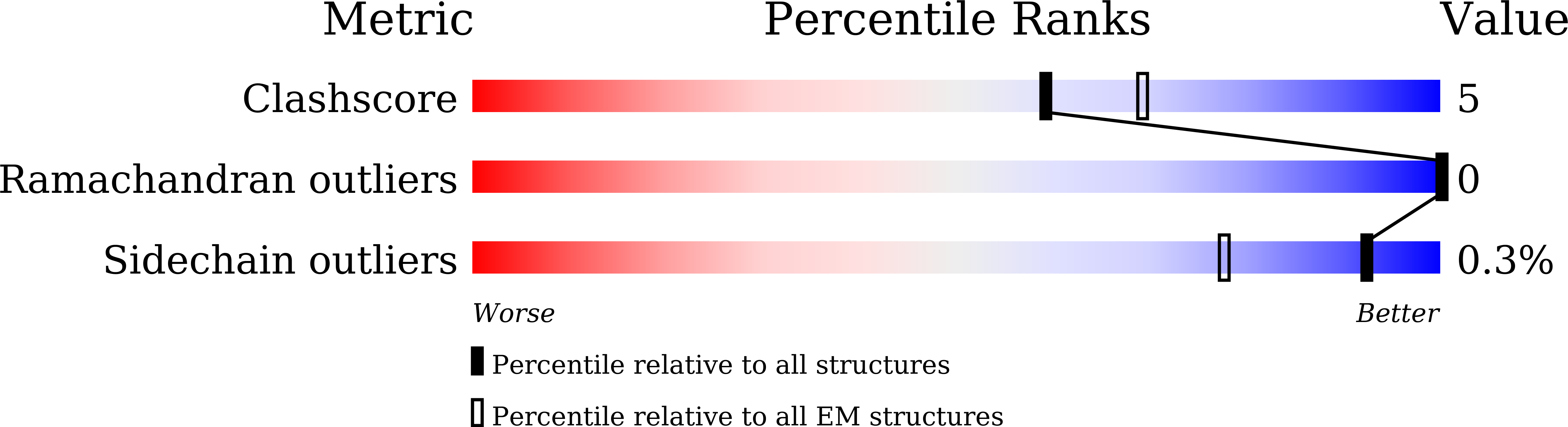Structural insights into the activation of human calcium-sensing receptor.
Chen, X., Wang, L., Cui, Q., Ding, Z., Han, L., Kou, Y., Zhang, W., Wang, H., Jia, X., Dai, M., Shi, Z., Li, Y., Li, X., Geng, Y.(2021) Elife 10
- PubMed: 34467854
- DOI: https://doi.org/10.7554/eLife.68578
- Primary Citation of Related Structures:
7E6T, 7E6U - PubMed Abstract:
Human calcium-sensing receptor (CaSR) is a G-protein-coupled receptor that maintains Ca 2+ homeostasis in serum. Here, we present the cryo-electron microscopy structures of the CaSR in the inactive and agonist+PAM bound states. Complemented with previously reported structures of CaSR, we show that in addition to the full inactive and active states, there are multiple intermediate states during the activation of CaSR. We used a negative allosteric nanobody to stabilize the CaSR in the fully inactive state and found a new binding site for Ca 2+ ion that acts as a composite agonist with L-amino acid to stabilize the closure of active Venus flytraps. Our data show that agonist binding leads to compaction of the dimer, proximity of the cysteine-rich domains, large-scale transitions of seven-transmembrane domains, and inter- and intrasubunit conformational changes of seven-transmembrane domains to accommodate downstream transducers. Our results reveal the structural basis for activation mechanisms of CaSR and clarify the mode of action of Ca 2+ ions and L-amino acid leading to the activation of the receptor.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.