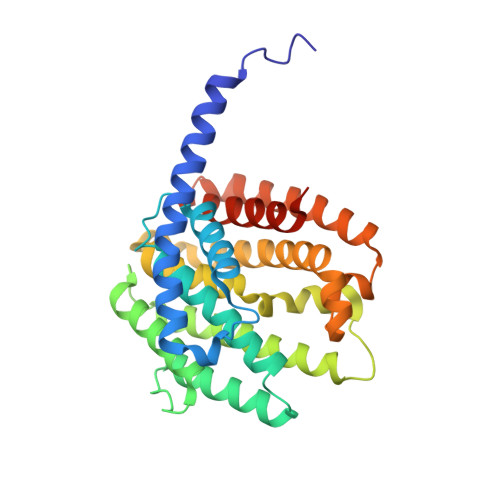
Functional characterization and structural bases of two class I diterpene synthases in pimarane-type diterpene biosynthesis
Xing, B., Yu, J., Chi, C., Ma, X., Xu, Q., Li, A., Ge, Y., Wang, Z., Liu, T., Jia, H., Yin, F., Guo, J., Huang, L., Yang, D., Ma, M.(2021) Commun Chem 4: 140