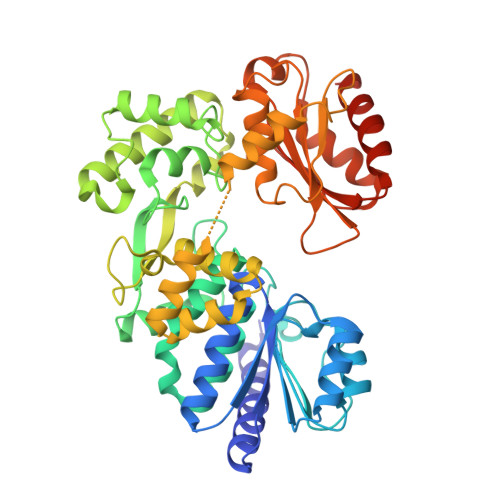
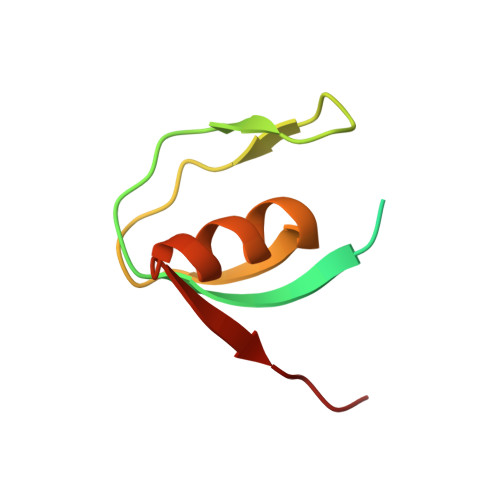
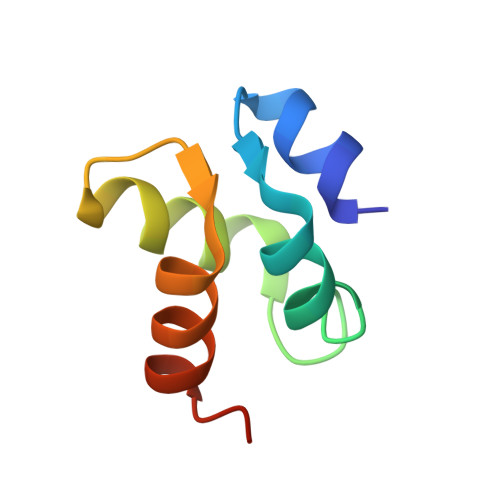
Family D DNA polymerase interacts with GINS to promote CMG-helicase in the archaeal replisome.
Oki, K., Nagata, M., Yamagami, T., Numata, T., Ishino, S., Oyama, T., Ishino, Y.(2022) Nucleic Acids Res 50: 3601-3615
- PubMed: 34568951
- DOI: https://doi.org/10.1093/nar/gkab799
- Primary Citation of Related Structures:
7E15 - PubMed Abstract:
Genomic DNA replication requires replisome assembly. We show here the molecular mechanism by which CMG (GAN-MCM-GINS)-like helicase cooperates with the family D DNA polymerase (PolD) in Thermococcus kodakarensis. The archaeal GINS contains two Gins51 subunits, the C-terminal domain of which (Gins51C) interacts with GAN. We discovered that Gins51C also interacts with the N-terminal domain of PolD's DP1 subunit (DP1N) to connect two PolDs in GINS. The two replicases in the replisome should be responsible for leading- and lagging-strand synthesis, respectively. Crystal structure analysis of the DP1N-Gins51C-GAN ternary complex was provided to understand the structural basis of the connection between the helicase and DNA polymerase. Site-directed mutagenesis analysis supported the interaction mode obtained from the crystal structure. Furthermore, the assembly of helicase and replicase identified in this study is also conserved in Eukarya. PolD enhances the parental strand unwinding via stimulation of ATPase activity of the CMG-complex. This is the first evidence of the functional connection between replicase and helicase in Archaea. These results suggest that the direct interaction of PolD with CMG-helicase is critical for synchronizing strand unwinding and nascent strand synthesis and possibly provide a functional machinery for the effective progression of the replication fork.
Organizational Affiliation:
Department of Bioscience and Biotechnology, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University, Fukuoka 819-0395, Japan.