Structural basis of an epitope tagging system derived from Haloarcula marismortui bacteriorhodopsin I D94N and its monoclonal antibody GD-26.
Pao, P.J., Hsu, M.F., Chiang, M.H., Chen, C.T., Lee, C.C., Wang, A.H.(2022) FEBS J 289: 730-747
- PubMed: 34499806
- DOI: https://doi.org/10.1111/febs.16184
- Primary Citation of Related Structures:
7DOH - PubMed Abstract:
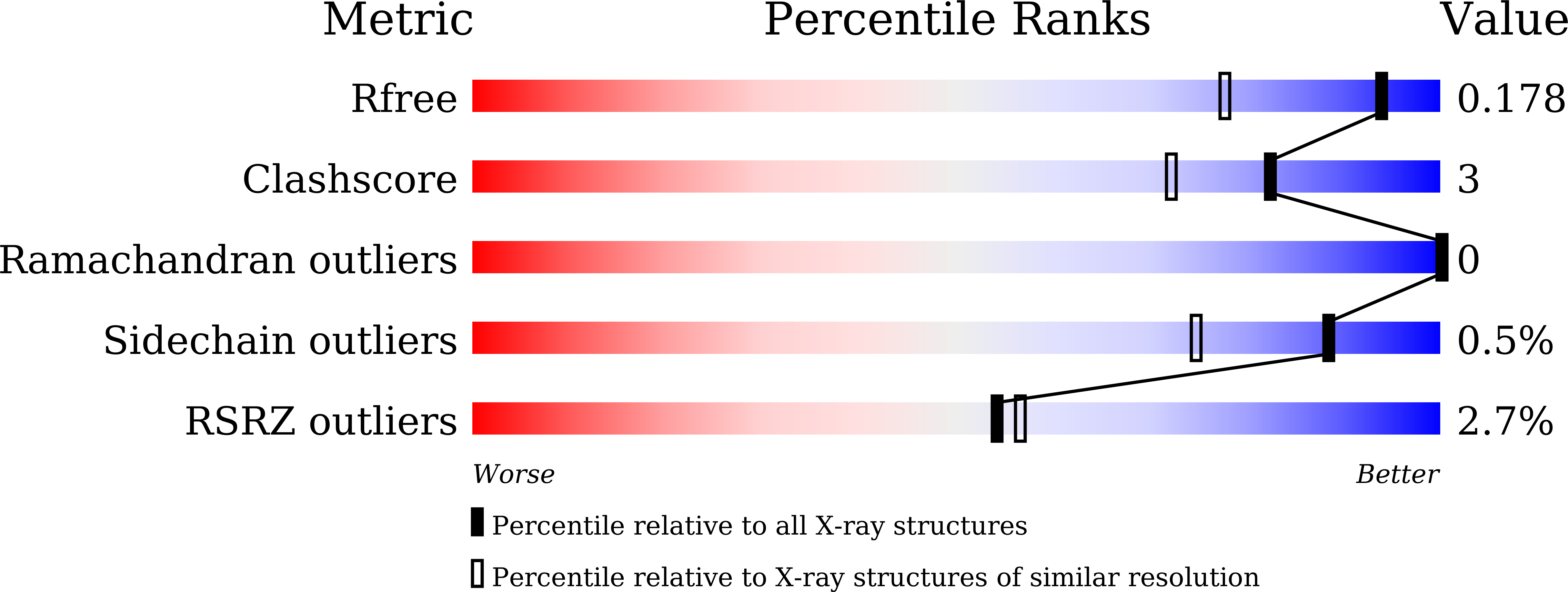
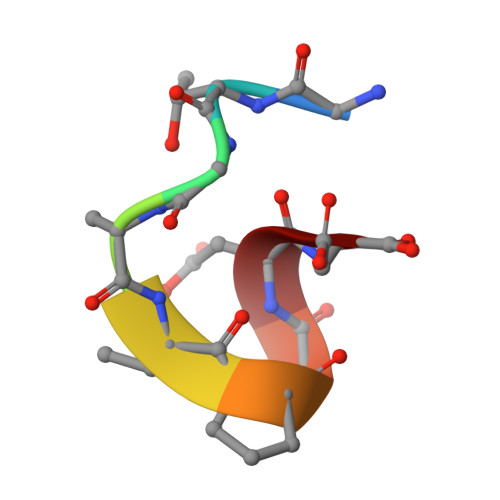
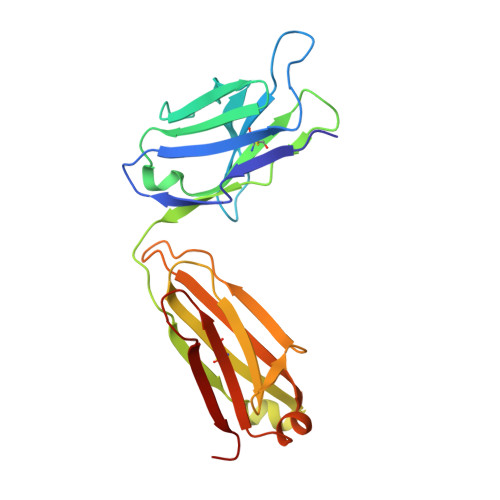
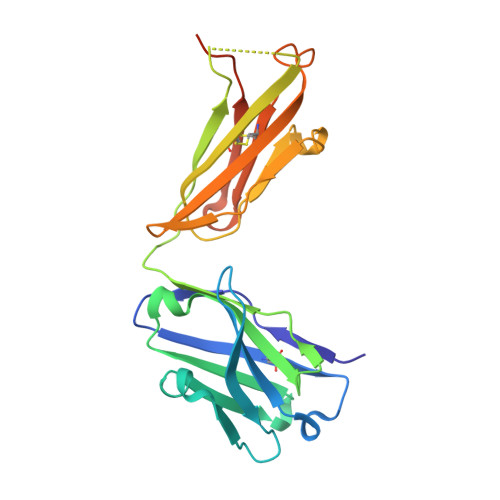
Specific antibody interactions with short peptides have made epitope tagging systems a vital tool employed in virtually all fields of biological research. Here, we present a novel epitope tagging system comprised of a monoclonal antibody named GD-26, which recognises the TD peptide (GTGATPADD) derived from Haloarcula marismortui bacteriorhodopsin I (HmBRI) D94N mutant. The crystal structure of the antigen-binding fragment (Fab) of GD-26 complexed with the TD peptide was determined to a resolution of 1.45 Å. The TD peptide was found to adopt a 3 10 helix conformation within the binding cleft, providing a characteristic peptide structure for recognition by GD-26 Fab. Based on the structure information, polar and nonpolar forces collectively contribute to the strong binding. Attempts to engineer the TD peptide show that the proline residue is crucial for the formation of the 3 10 helix in order to fit into the binding cleft. Isothermal calorimetry (ITC) reported a dissociation constant K D of 12 ± 2.8 nm, indicating a strong interaction between the TD peptide and GD-26 Fab. High specificity of GD-26 IgG to the TD peptide was demonstrated by western blotting, ELISA and immunofluorescence as only TD-tagged proteins were detected, suggesting the effectiveness of the GD-26/TD peptide tagging system. In addition to already-existing epitope tags such as the FLAG tag and the ALFA tag adopting either extended or α-helix conformations, the unique 3 10 helix conformation of the TD peptide together with the corresponding monoclonal antibody GD-26 offers a novel tagging option for research.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.