Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs.
Niu, M., Xu, S., Yang, J., Yao, D., Li, N., Yan, J., Zhong, G., Song, G.(2021) J Biol Chem 296: 100776-100776
- PubMed: 33992645
- DOI: https://doi.org/10.1016/j.jbc.2021.100776
- Primary Citation of Related Structures:
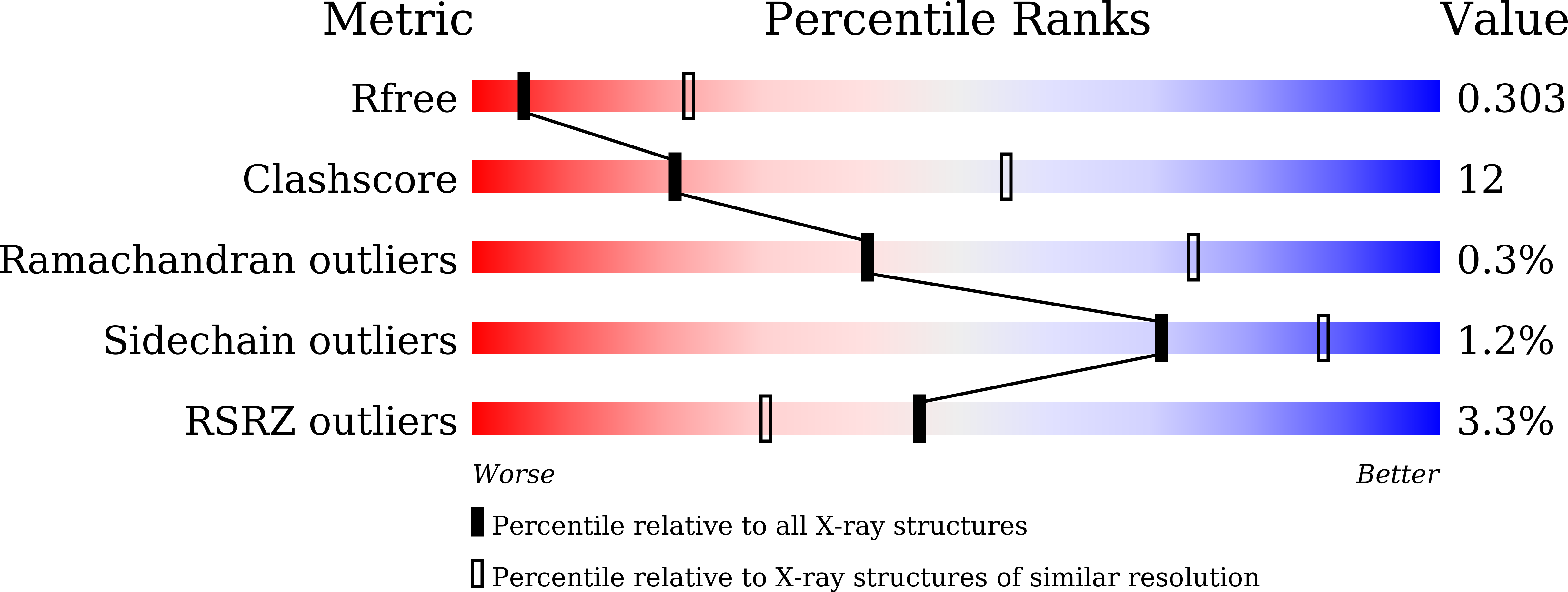
7DO4 - PubMed Abstract:
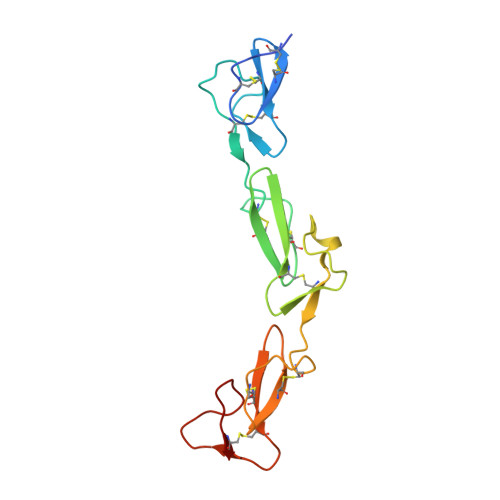
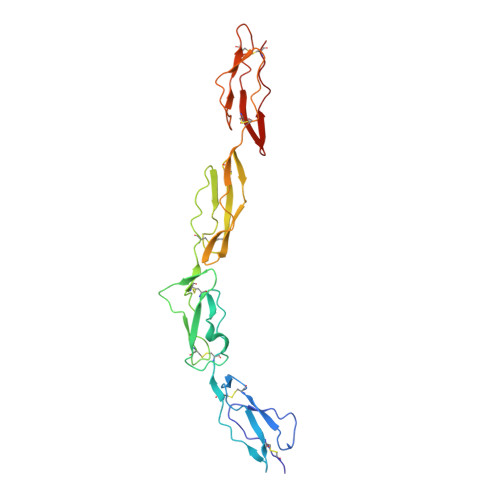
The adhesion G protein-coupled receptor CD97 and its ligand complement decay-accelerating factor CD55 are important binding partners in the human immune system. Dysfunction in this binding has been linked to immune disorders such as multiple sclerosis and rheumatoid arthritis, as well as various cancers. Previous literatures have indicated that the CD97 includes 3 to 5 epidermal growth factor (EGF) domains at its N terminus and these EGF domains can bind to the N-terminal short consensus repeat (SCR) domains of CD55. However, the details of this interaction remain elusive, especially why the CD55 binds with the highest affinity to the shortest isoform of CD97 (EGF 1,2,5 ). Herein, we designed a chimeric expression construct with the EGF 1,2,5 domains of CD97 and the SCR 1-4 domains of CD55 connected by a flexible linker and determined the complex structure by crystallography. Our data reveal that the two proteins adopt an overall antiparallel binding mode involving the SCR 1-3 domains of CD55 and all three EGF domains of CD97. Mutagenesis data confirmed the importance of EGF 5 in the interaction and explained the binding specificity between CD55 and CD97. The architecture of CD55-CD97 binding mode together with kinetics suggests a force-resisting shearing stretch geometry when forces applied to the C termini of both proteins in the circulating environment. The potential of the CD55-CD97 complex to withstand tensile force may provide a basis for the mechanosensing mechanism for activation of adhesion G protein-coupled receptors.
Organizational Affiliation:
Shanghai Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai, China.