Design of complicated all-alpha protein structures
Sakuma, K., Kobayashi, N., Sugiki, T., Nagashima, T., Fujiwara, T., Suzuki, K., Kobayashi, N., Murata, T., Kosugi, T., Koga, R., Koga, N.(2024) Nat Struct Mol Biol
Experimental Data Snapshot
(2024) Nat Struct Mol Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| de novo designed protein | 97 | synthetic construct | Mutation(s): 0 | 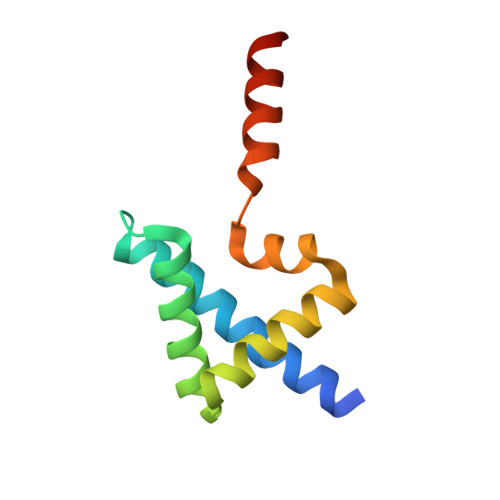 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL (Subject of Investigation/LOI) Query on GOL | C [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.98 | α = 90 |
| b = 33.66 | β = 93.11 |
| c = 58.38 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XDS | data scaling |
| MOLREP | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |