Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Wang, W., Zhang, Y., Zhao, G., Wang, H.(2021) Inorg Chem 60: 7207-7216
- PubMed: 33852289
- DOI: https://doi.org/10.1021/acs.inorgchem.1c00265
- Primary Citation of Related Structures:
7DIE - PubMed Abstract:
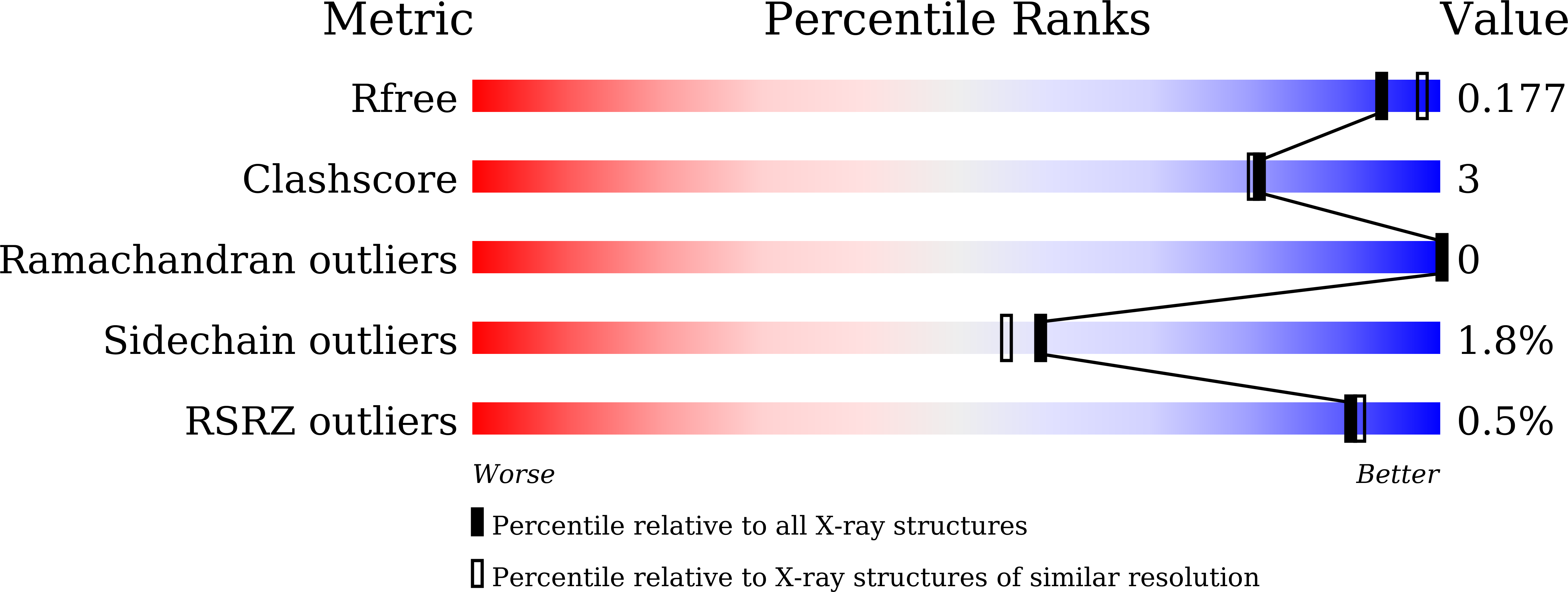
Here, we present a 1.9 Å resolution crystal structure of Mycoplasma Penetrans ferritin, which reveals that its ferroxidase center is located on the inner surface of ferritin but not buried within the four-helix of each subunit. Such a ferroxidase center exhibits a lower iron oxidation activity as compared to the reported ferritin. More importantly, we found that Fe 2+ enters into the center via the rarely reported B-channels rather than the normal 3- or 4-fold channels. All these findings may provide the structural bases to explore the new iron oxidation mechanism adopted by this special ferritin, which is beneficial for understanding the relationship between the structure and function of ferritin.
Organizational Affiliation:
Institute of Molecular Science, Shanxi University, Taiyuan 030006, China.