Structure Basis for Shaping the Nse4 Protein by the Nse1 and Nse3 Dimer within the Smc5/6 Complex.
Jo, A., Li, S., Shin, J.W., Zhao, X., Cho, Y.(2021) J Mol Biol 433: 166910-166910
- PubMed: 33676928
- DOI: https://doi.org/10.1016/j.jmb.2021.166910
- Primary Citation of Related Structures:
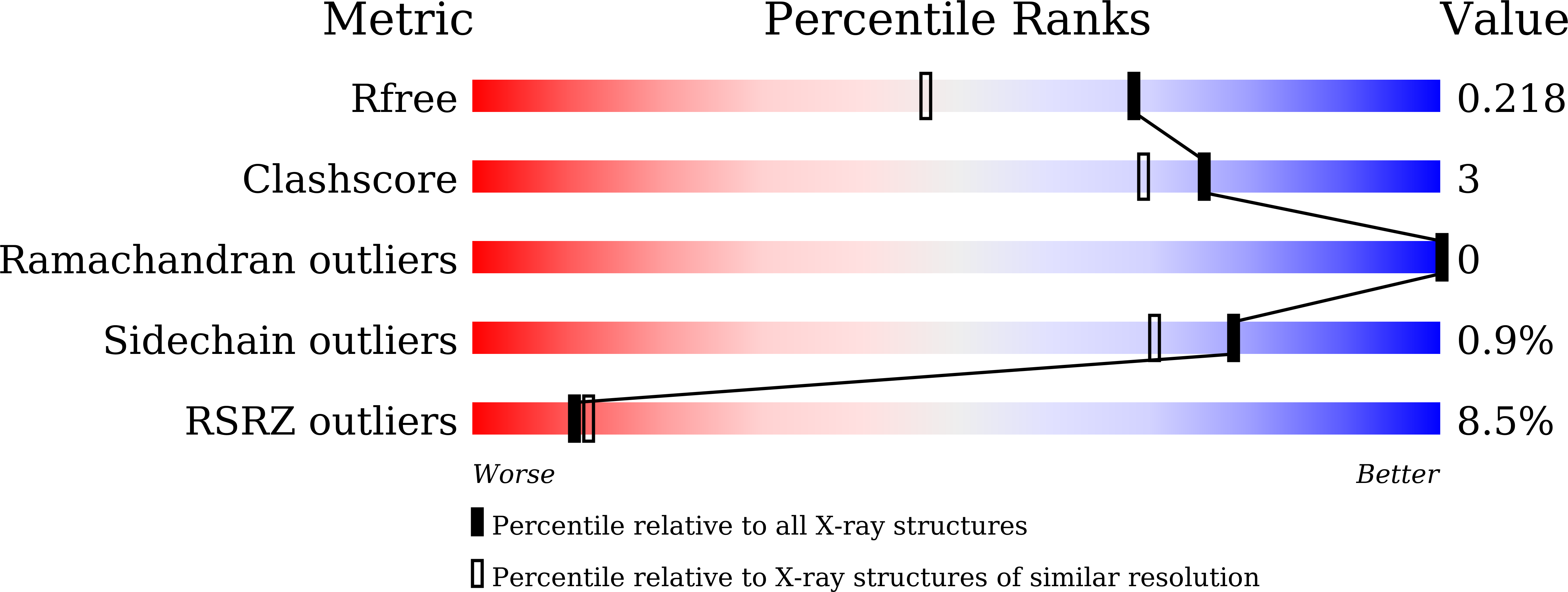
7DG2 - PubMed Abstract:
The Smc5/6 complex facilitates chromosome replication and DNA break repair. Within this complex, a subcomplex composed of Nse1, Nse3 and Nse4 is thought to play multiple roles through DNA binding and regulating ATP-dependent activities of the complex. However, how the Nse1-Nse3-Nse4 subcomplex carries out these multiple functions remain unclear. To address this question, we determine the crystal structure of the Xenopus laevis Nse1-Nse3-Nse4 subcomplex at 1.7 Å resolution and examine how it interacts with DNA. Our structural analyses show that the Nse1-Nse3 dimer adopts a closed conformation and forms three interfaces with a segment of Nse4, forcing it into a Z-shaped conformation. The Nse1-Nse3-Nse4 structure provides an explanation for how the lung disease immunodeficiency and chromosome breakage syndrome-causing mutations could dislodge Nse4 from Nse1-Nse3. Our DNA binding and mutational analyses reveal that the N-terminal and the middle region of Nse4 contribute to DNA interaction and cell viability. Integrating our data with previous crosslink mass spectrometry data, we propose potential roles of the Nse1-Nse3-Nse4 complex in binding DNA within the Smc5/6 complex.
Organizational Affiliation:
Department of Life Science, Pohang University of Science and Technology, Pohang, Republic of Korea.